I have two sets of geolocation data as lat/long in a csv file. The first two columns (0 and 1) comprise the latitude and longitude of the first set, and the next two columns (2 and 3) comprise the second set. The first set contains about 100 entries and the second set contains about 75. I want to plot both sets onto the same graph. I've loaded the csv file into Python using numpy.loadtxt.
The first set loads without any issue, but when I try to load the second set I get this error:
ValueError: could not convert string to float: ' '
After several hours of frustration, I finally determined that Python is reading blank csv cells in the second data set so that it will be equal with the first data set. This is literally the dumbest reason for anything I've ever heard of, because it is reading blank cells and expecting them to contain information, and then complains that there is no information.
Why is Python reading blank cells and how can I make it stop doing that?
To recreate the error, fill two columns with data and the next two with less data. If your first two columns have 10 entries, the next two have no more than 9. Save as a csv file and run the following code:
import numpy as np
import matplotlib.pyplot as plt
lat1 = np.loadtxt(r'C:\Users\XXXXX\Desktop\latlong.csv', usecols=0, delimiter=',')
long1 = np.loadtxt(r'C:\Users\XXXXX\Desktop\latlong.csv', usecols=1, delimiter=',')
lat2 = np.loadtxt(r'C:\Users\XXXXX\Desktop\latlong.csv', usecols=2, delimiter=',')
long2 = np.loadtxt(r'C:\Users\XXXXX\Desktop\latlong.csv', usecols=3, delimiter=',')
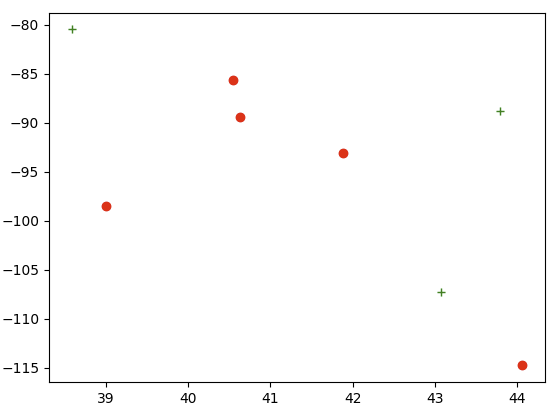
plt.plot(lat1, long1, 'o')
plt.plot(lat2, long2, ' ')
You will get this error:
Traceback (most recent call last):
File "C:\Users\XXXXX\AppData\Local\Programs\Spyder\pkgs\spyder_kernels\py3compat.py", line 356, in compat_exec
exec(code, globals, locals)
File "c:\users\XXXXX\XXXXX\XXXXX\XXXXX\XXXXX\untitled0.py", line 11, in <module>
lat1 = np.loadtxt(r'C:\Users\XXXXX\Desktop\latlong.csv', usecols=0, delimiter=',')
File "C:\Users\XXXXX\AppData\Local\Programs\Spyder\pkgs\numpy\lib\npyio.py", line 1163, in loadtxt
chunk.append(packer(convert_row(words)))
File "C:\Users\XXXXX\AppData\Local\Programs\Spyder\pkgs\numpy\lib\npyio.py", line 1142, in convert_row
return [*map(_conv, vals)]
File "C:\Users\XXXXX\AppData\Local\Programs\Spyder\pkgs\numpy\lib\npyio.py", line 725, in _floatconv
return float(x) # The fastest path.
ValueError: could not convert string to float: ''
Here is a sample of the data I'm trying to use:
41.878 -93.0977 43.78444 -88.7879
44.062 -114.742 38.58763 -80.4549
40.63313 -89.3985 43.07597 -107.29
40.55122 -85.6024
39.0119 -98.4842
The first two columns have two more lat/long entries than the second two columns. For reasons unknown to me, Python is reading an extra 4 blank cells in the second two columns, and then tries to use the blank cells as plotting data, giving me a ValueError. Why is Python reading blank data and trying to plot it? What is the solution to this?
CodePudding user response:
CodePudding user response:
With your sample:
In [1]: txt='''41.878 -93.0977 43.78444 -88.7879
...: 44.062 -114.742 38.58763 -80.4549
...: 40.63313 -89.3985 43.07597 -107.29
...: 40.55122 -85.6024
...: 39.0119 -98.4842'''
I get an error on the lat1 line, as you do, but for a different string:
In [2]: lat1 = np.loadtxt(txt.splitlines(), usecols=0, delimiter=',')
Traceback (most recent call last):
Input In [2] in <cell line: 1>
lat1 = np.loadtxt(txt.splitlines(), usecols=0, delimiter=',')
File /usr/local/lib/python3.10/dist-packages/numpy/lib/npyio.py:1313 in loadtxt
arr = _read(fname, dtype=dtype, comment=comment, delimiter=delimiter,
File /usr/local/lib/python3.10/dist-packages/numpy/lib/npyio.py:979 in _read
arr = _load_from_filelike(
ValueError: could not convert string '41.878 -93.0977 43.78444 -88.7879' to float64 at row 0, column 1.
There is a comma delimiter, so it tries to convert the whole line to float, with an obvious failure.
Letting it use the default white space delimiter:
In [3]: lat1 = np.loadtxt(txt.splitlines(), usecols=0)
In [4]: lat1
Out[4]: array([41.878 , 44.062 , 40.63313, 40.55122, 39.0119 ])
In [5]: np.loadtxt(txt.splitlines(), usecols=1)
Out[5]: array([ -93.0977, -114.742 , -89.3985, -85.6024, -98.4842])
But it fails when we ask it to load column 2 on the shorter lines:
In [6]: np.loadtxt(txt.splitlines(), usecols=2)
Traceback (most recent call last):
Input In [6] in <cell line: 1>
np.loadtxt(txt.splitlines(), usecols=2)
File /usr/local/lib/python3.10/dist-packages/numpy/lib/npyio.py:1313 in loadtxt
arr = _read(fname, dtype=dtype, comment=comment, delimiter=delimiter,
File /usr/local/lib/python3.10/dist-packages/numpy/lib/npyio.py:979 in _read
arr = _load_from_filelike(
ValueError: invalid column index 2 at row 4 with 2 columns
Trying to read the whole sample, both loadtxt and genfromtxt have problems with the short lines:
In [7]: np.loadtxt(txt.splitlines())
Traceback (most recent call last):
Input In [7] in <cell line: 1>
np.loadtxt(txt.splitlines())
File /usr/local/lib/python3.10/dist-packages/numpy/lib/npyio.py:1313 in loadtxt
arr = _read(fname, dtype=dtype, comment=comment, delimiter=delimiter,
File /usr/local/lib/python3.10/dist-packages/numpy/lib/npyio.py:979 in _read
arr = _load_from_filelike(
ValueError: the number of columns changed from 4 to 2 at row 4; use `usecols` to select a subset and avoid this error
In [8]: np.genfromtxt(txt.splitlines())
Traceback (most recent call last):
Input In [8] in <cell line: 1>
np.genfromtxt(txt.splitlines())
File /usr/local/lib/python3.10/dist-packages/numpy/lib/npyio.py:2266 in genfromtxt
raise ValueError(errmsg)
ValueError: Some errors were detected !
Line #4 (got 2 columns instead of 4)
Line #5 (got 2 columns instead of 4)
(I'm using the latest numpy version, which has a reword loadtxt, so its error messages may be different.)
Changing the text to use comma, including markers for the missing fields, genfromtxt works fine:
In [9]: txt='''41.878, -93.0977, 43.78444, -88.7879
...: 44.062 , -114.742 , 38.58763 , -80.4549
...: 40.63313 , -89.3985, 43.07597, -107.29
...: 40.55122 , -85.6024,,
...: 39.0119 , -98.4842,,'''
In [12]: np.genfromtxt(txt.splitlines(), delimiter=',')
Out[12]:
array([[ 41.878 , -93.0977 , 43.78444, -88.7879 ],
[ 44.062 , -114.742 , 38.58763, -80.4549 ],
[ 40.63313, -89.3985 , 43.07597, -107.29 ],
[ 40.55122, -85.6024 , nan, nan],
[ 39.0119 , -98.4842 , nan, nan]])