I have a data frame where some columns have entries in every row, and other variables only have entries in some rows, and NAs in the rest. A reproducible example is below:
df <- data.frame(id = 1:5,
var1 = c("Agree", "Strongly Agree", "Disagree", "Disagree", "Strongly Agree"),
var2 = c("Strongly Agree", NA, "Agree", NA, NA))
I would like to summarize this data frame and calculate the percentage of responses that fall into each category for each column, i.e. the percentage of "Agree" in var1, and the percentage of "Strongly Agree" in var2, among others. I would like the denominator of the percentages, so to speak, not to include the rows with NA, i.e. have different Ns depending on the row. This is what I have tried thus far:
df %>%
filter(filter(!is.na(var1)) %>%
group_by(var1) %>%
summarize(frequency = n()/count(filter(df, !is.na(var1))))
This produces a list where the first column is populated with all of the different possible levels, but the second column is a data.frame that in turn includes the frequency values. The problem with this is that I would like to pipe this into ggplot to make a bar graph of the percentages, but since the second column is a data.frame, ggplot will not take the data, throwing the error Error in is.finite(x) : default method not implemented for type 'list'.
Is there a way to make the output of the summarize() function a non-nested data.frame, or alternatively, is there a way to force ggplot to accept the output as shown here? Any assistance would be appreciated!
CodePudding user response:
I think you can achieve what you want by reshaping the data from wide to long format using tidyr::pivot_longer.
library(dplyr)
library(tidyr)
library(ggplot2)
df %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
count(name, value) %>%
group_by(name) %>%
mutate(frequency = n/sum(n))
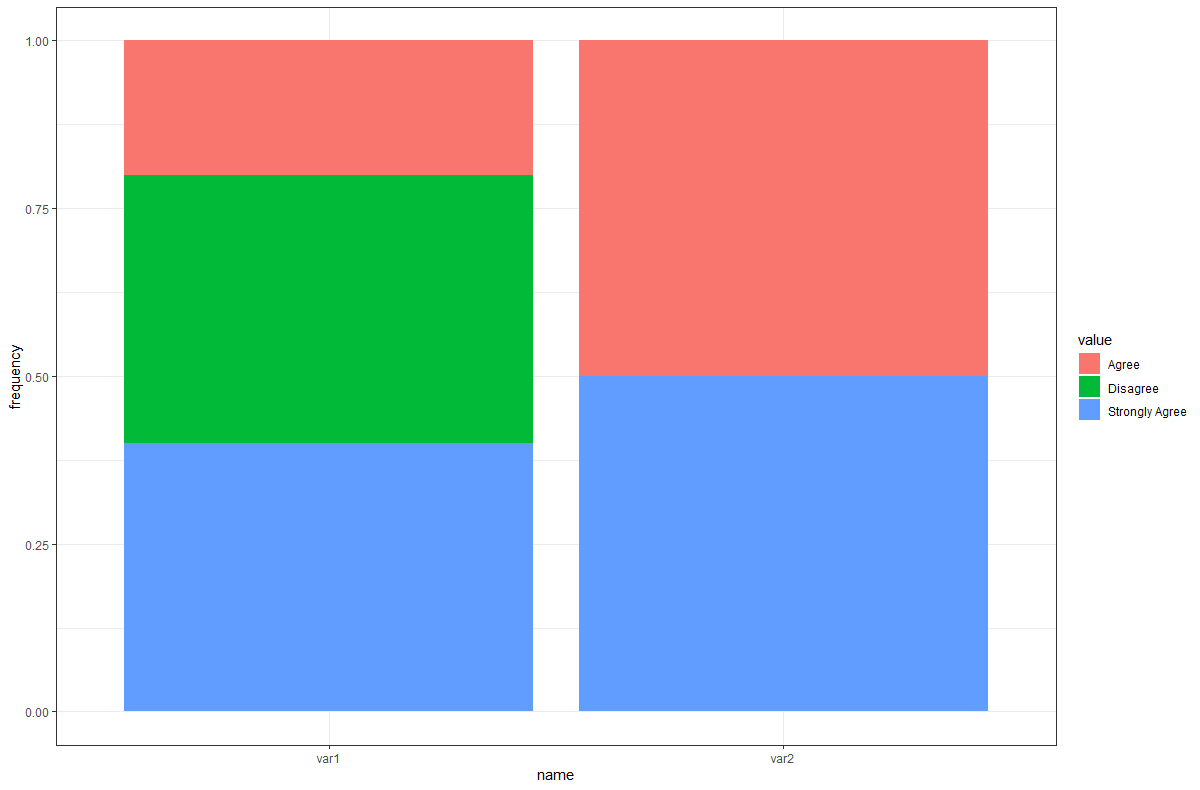
Result:
name value n frequency
<chr> <chr> <int> <dbl>
1 var1 Agree 1 0.2
2 var1 Disagree 2 0.4
3 var1 Strongly Agree 2 0.4
4 var2 Agree 1 0.5
5 var2 Strongly Agree 1 0.5
And you can pipe into ggplot.
df %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
count(name, value) %>%
group_by(name) %>%
mutate(frequency = n/sum(n)) %>%
ggplot(aes(name, frequency))
geom_col(aes(fill = value))
Result: