Here is a sample of my data:
> dput(x)
structure(list(guide_id = c("ALPI_g06__safe_g06", "SAMD7_g05__safe_g05",
"GK_g03__GK2_g03", "safe_g03__CDK1_g03", "safe_g03__GOLPH3L_g03",
"UBE2V2_g06__safe_g06", "PIK3R2_g03__safe_g03", "safe_g06__BUB3_g06",
"ATG5_g04__PTEN_g06", "PTEN_g05__MCM9_g05", "PTEN_g01__LRP1B_g03",
"ORC3_g02__PTEN_g06", "CDKN2A_g05__PTMA_g05", "KIF4A_g03__safe_g03",
"safe_g06__CCNYL1_g06", "CSTF2T_g06__safe_g06", "HDAC2_g03__HDAC1_g01",
"safe_g05__NDST4_g05", "PTEN_g03__IRF2_g01", "METTL7A_g05__METTL7B_g05",
"PTEN_g05__PPP3CC_g05", "CBLB_g06__CBL_g06", "MAP2K6_g02__MAP2K3_g06",
"CAB39L_g05__CAB39_g01", "IFNA8_g06__PRKN_g02", "safe_g01__RPL23_g01",
"LIN28A_g02__safe_g02", "KAT6A_g06__safe_g06", "CHSY1_g02__safe_g02",
"safe_g06__NET1_g06", "ADH1A_g01__ADH1B_g05", "safe_g05__GRIA4_g05",
"NIPSNAP3B_g04__NIPSNAP3A_g04", "MX1_g06__safe_g06", "safe_g01__safe_g04",
"safe_g04__UBTD1_g04", "safe_g03__FARSB_g03", "HIPK2_g04__HIPK1_g04",
"PASK_g04__CDKN2A_g02", "DNMT3B_g06__safe_g06", "CPEB4_g02__CPEB3_g06",
"safe_g05__GIT2_g05", "HEATR5A_g05__HEATR5B_g05", "CDKN2A_g05__PTK2B_g01",
"CDKN2A_g01__CYSLTR2_g01", "MTAP_g01__PDE4D_g01", "PTEN_g03__FIG4_g01",
"CELA3B_g05__safe_g05", "PLS3_g02__LCP1_g06", "PTEN_g01__PPP3CC_g03",
"safe_g01__PDE6H_g01", "ADH1B_g05__ADH1C_g03", "ATP2B4_g04__ATP2B3_g06",
"CDKN2A_g05__TEX15_g03", "ORC3_g03__safe_g03", "PTEN_g03__CHMP7_g05",
"FDFT1_g06__safe_g06", "STAR_g06__safe_g06", "IRF2_g02__CDKN2A_g02",
"NAA38_g05__NUP205_g03", "STK24_g02__STK25_g06", "safe_g05__LDB1_g05",
"AIMP2_g01__MRPL1_g03", "ROCK1_g03__safe_g03", "AGPAT2_g06__safe_g06",
"ALPG_g01__ALPI_g05", "CAD_g05__safe_g05", "ATAD1_g06__KLHL9_g02",
"CDKN2A_g05__KLHL9_g01", "PTEN_g05__PLAA_g01", "ZNF561_g05__ZNF562_g05",
"safe_g06__OR5I1_g06", "CCAR2_g02__PTEN_g02", "safe_g06__COPS7A_g06",
"MYH7_g01__MYH6_g01", "ITSN2_g02__safe_g02", "PNPLA2_g01__safe_g01",
"PGM2_g05__PGM2L1_g05", "CCAR2_g04__CDKN2A_g04", "ELMO3_g04__ELMO2_g02",
"DUSP16_g05__safe_g05", "MYH7_g01__MYH3_g03", "safe_g01__MUCL3_g01",
"safe_g03__CIAO3_g03", "safe_g06__UTY_g06", "PRKCG_g04__PRKCB_g02",
"safe_g03__PUF60_g03", "safe_g05__PUF60_g05", "PPP2R1A_g05__DDX3X_g05",
"MTAP_g01__WRN_g01", "IFNA2_g02__RB1_g06", "SNRPA1_g02__safe_g02",
"STK3_g03__STK4_g05", "RB1_g02__PTEN_g02", "PRKCZ_g02__PRKCI_g04",
"KMT2A_g05__KMT2B_g05", "DDX39A_g01__DDX39B_g05", "safe_g06__UBP1_g06",
"safe_g03__HACE1_g03", "PER2_g02__CDKN2B_g02"), orientation = c("gene__safe",
"gene__safe", "gene__gene", "safe__gene", "safe__gene", "gene__safe",
"gene__safe", "safe__gene", "gene__gene", "gene__gene", "gene__gene",
"gene__gene", "gene__gene", "gene__safe", "safe__gene", "gene__safe",
"gene__gene", "safe__gene", "gene__gene", "gene__gene", "gene__gene",
"gene__gene", "gene__gene", "gene__gene", "gene__gene", "safe__gene",
"gene__safe", "gene__safe", "gene__safe", "safe__gene", "gene__gene",
"safe__gene", "gene__gene", "gene__safe", "safe__safe", "safe__gene",
"safe__gene", "gene__gene", "gene__gene", "gene__safe", "gene__gene",
"safe__gene", "gene__gene", "gene__gene", "gene__gene", "gene__gene",
"gene__gene", "gene__safe", "gene__gene", "gene__gene", "safe__gene",
"gene__gene", "gene__gene", "gene__gene", "gene__safe", "gene__gene",
"gene__safe", "gene__safe", "gene__gene", "gene__gene", "gene__gene",
"safe__gene", "gene__gene", "gene__safe", "gene__safe", "gene__gene",
"gene__safe", "gene__gene", "gene__gene", "gene__gene", "gene__gene",
"safe__gene", "gene__gene", "safe__gene", "gene__gene", "gene__safe",
"gene__safe", "gene__gene", "gene__gene", "gene__gene", "gene__safe",
"gene__gene", "safe__gene", "safe__gene", "safe__gene", "gene__gene",
"safe__gene", "safe__gene", "gene__gene", "gene__gene", "gene__gene",
"gene__safe", "gene__gene", "gene__gene", "gene__gene", "gene__gene",
"gene__gene", "safe__gene", "safe__gene", "gene__gene"), method = structure(c(1L,
1L, 3L, 2L, 1L, 2L, 3L, 2L, 2L, 1L, 1L, 2L, 1L, 3L, 2L, 3L, 1L,
3L, 3L, 1L, 1L, 3L, 1L, 2L, 2L, 1L, 3L, 3L, 3L, 1L, 1L, 3L, 3L,
1L, 3L, 3L, 2L, 3L, 2L, 2L, 1L, 2L, 2L, 1L, 3L, 1L, 1L, 2L, 2L,
2L, 2L, 3L, 1L, 3L, 2L, 1L, 1L, 1L, 2L, 3L, 2L, 3L, 2L, 2L, 2L,
2L, 1L, 1L, 3L, 1L, 2L, 1L, 3L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 2L, 3L, 3L, 3L, 3L, 3L, 1L, 3L,
3L, 3L, 2L), .Label = c("exact_trimmed", "TinQ_trimmed", "TinQ_untrimmed"
), class = "factor"), sample_id = structure(c(2L, 9L, 10L, 4L,
3L, 1L, 1L, 7L, 6L, 2L, 8L, 10L, 3L, 8L, 3L, 3L, 2L, 10L, 7L,
5L, 7L, 5L, 8L, 4L, 8L, 9L, 4L, 6L, 5L, 9L, 8L, 10L, 6L, 4L,
7L, 1L, 2L, 10L, 1L, 3L, 5L, 5L, 5L, 6L, 10L, 2L, 6L, 6L, 5L,
4L, 1L, 5L, 6L, 6L, 10L, 2L, 6L, 4L, 3L, 4L, 1L, 9L, 6L, 2L,
4L, 5L, 7L, 6L, 8L, 1L, 10L, 7L, 3L, 2L, 6L, 3L, 6L, 8L, 10L,
2L, 10L, 7L, 1L, 3L, 1L, 4L, 3L, 1L, 5L, 10L, 5L, 7L, 6L, 7L,
1L, 1L, 3L, 5L, 3L, 2L), .Label = c("hiseq_pDNA", "miseq_pDNA",
"A01", "A02", "A03", "A06", "A10", "A11", "A12", "B03"), class = "factor"),
normalised_count = c(269.032403623316, 272.950841923723,
165.681100384761, 166.943692848127, 40.8506826249191, 424.743797348522,
329.747668529917, 4.29849062800088, 34.5651047326788, 250.478444752743,
704.835258433083, 194.602213349721, 592.334898061327, 28.6264448807693,
29.2362728590107, 117.639710460663, 214.607457602967, 203.778893521364,
63.7658744401034, 173.057914568954, 324.701368976682, 0,
7.8141381201007, 622.344192683017, 51.5733115926646, 609.972912684359,
76.5704657212621, 397.17161185068, 445.473347378776, 90.3663574347198,
134.403175665732, 27.465850865923, 138.146647600236, 290.098876096745,
5047.7462678544, 647.537959958331, 210.278200533167, 289.277429281415,
924.036343527445, 46.0571421751539, 40.6932047177645, 436.445800049869,
45.1649854559803, 584.314865719095, 16.8339085952431, 142.24701800773,
117.68595182793, 2.4689360523342, 183.343010266851, 331.150603846284,
246.655119587007, 860.563357122891, 19.7514884186736, 2.46690442143279,
22.1642612015627, 933.264131189848, 212.328500500741, 77.7246045391279,
130.962482532829, 114.308766683884, 761.593699471077, 1453.74759034584,
829.562513584292, 259.755424188029, 528.198897044073, 153.382079320804,
779.018763043852, 83.1208470952515, 1408.94156894986, 484.106689935693,
10.6388453767501, 56.2110312892423, 186.062807361253, 247.386118274314,
344.828068642677, 72.890433703287, 1371.08248772959, 95.8534276065686,
587.352921841412, 577.646586170522, 115.254158248126, 3.63718437753921,
365.841082223266, 140.17391096786, 445.451783134745, 330.055891106296,
315.992045010404, 494.391553235232, 317.237790008958, 297.887670549003,
521.878505254173, 331.053918077635, 153.770375602644, 209.799638701895,
274.559774215287, 191.43382415708, 206.069560840957, 58.5326063255964,
303.702517821916, 859.048295707554), ref_normalised_count = c(271.044525068558,
240.672812582099, 167.403279421046, 280.24807430688, 144.495723041642,
424.743797348522, 329.747668529917, 121.947027407755, 630.443122825,
235.610860501022, 508.035917955329, 221.80553664354, 599.151055414708,
363.320304237984, 212.141809943303, 246.965827057972, 251.717071668084,
162.80428822816, 354.582220971501, 442.230540901332, 280.708251768796,
96.1189159313148, 222.725891567372, 407.717231257628, 354.796823137281,
194.655066390493, 229.949559644294, 177.980959164683, 339.405550034978,
192.354179080912, 580.28377947615, 250.185120892992, 206.494704560576,
376.425163847336, 602.927745387338, 647.537959958331, 227.787843648449,
362.860405118696, 924.036343527445, 113.20365563135, 143.57536811781,
241.593167505931, 153.699272279964, 468.460656230547, 224.430770212831,
167.504596137445, 142.655013193978, 173.026725680438, 274.725944763887,
292.672865778613, 246.655119587007, 547.279951953419, 298.194995321606,
203.735309844844, 174.407258066186, 847.64688484938, 362.619839989854,
224.10642395312, 259.079911058741, 720.661919925217, 761.593699471077,
698.126863080076, 76.3894586780657, 294.053398164361, 391.611020090566,
358.938420294526, 156.46033705146, 410.478296029124, 848.053975968156,
484.106689935693, 123.327559793504, 169.805483447026, 542.221061641245,
264.602040601734, 372.743744152007, 252.637426591916, 212.601987405219,
229.628553496113, 387.469422933321, 539.32798536562, 265.06221806365,
176.24796791385, 365.841082223266, 394.832262323978, 445.451783134745,
364.000372375602, 627.221880591588, 494.391553235232, 413.44930824044,
559.575793689927, 864.610344262545, 253.404414728012, 87.3808326648316,
389.994453156722, 274.559774215287, 191.43382415708, 361.48070776083,
229.949559644294, 252.484616489435, 832.921206068066)), row.names = c(NA,
-100L), class = c("tbl_df", "tbl", "data.frame"))
I am trying to plot as:
x %>% ggplot(aes(x = ref_normalised_count, y = normalised_count, col = orientation))
geom_point(size = 0.3)
theme_manuscript(base_size = 8)
labs(title = 'Normalised counts')
facet_grid(. ~ method sample_id)
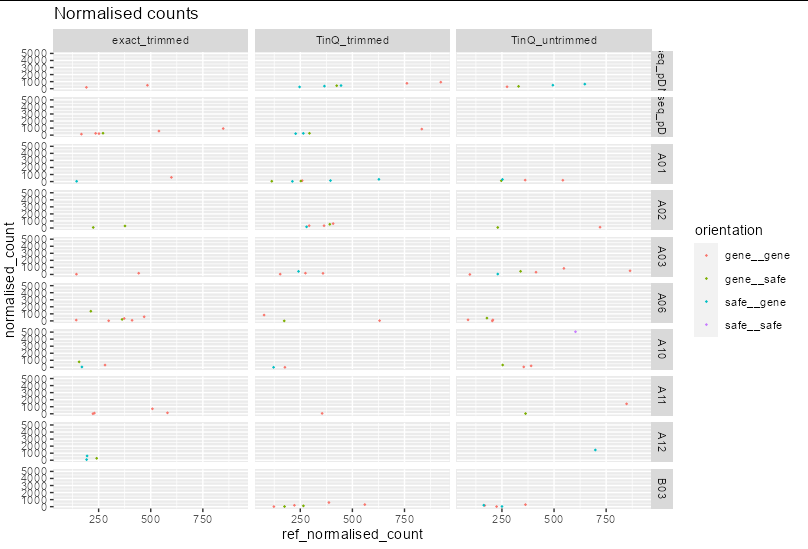
This gives me the plots I would like which is the normalised count vs the reference normalised count for each sample by each method. However, rather than having all in one row, I would like to have method as columns and samples as rows to form a 10 rows x 3 columns matrix of plots. How is this possible?
CodePudding user response:
You need to put the two faceting variables on opposite sides of the tilde in the formula:
x %>%
ggplot(aes(x = ref_normalised_count, y = normalised_count, col = orientation))
geom_point(size = 0.3)
labs(title = 'Normalised counts')
facet_grid(sample_id ~ method)