ggplot is refusing to cooperate with color assignments. It continues to assign colors based on alphabetical order, even after I've assigned Factor levels. Please note: I have simplified my code here. This is actually a part of a much more complicated function that creates many plots from distinct datasets and is the reasoning behind the use of [] to call appropriate coloring.
Thanks for any help!
Example Data
LUT <- data.frame(
Mutation = c("Wildtype", "Deletion"),
ConditionID = c("Control_Rep1","Treatment_Rep1"),
Treatment = c("Control", "Treatment")
)
data <- data.frame(
bp = c(1:10),
cAvgCPM = c(10.5, 10.4, 10.4, 10.4, 10.3, 12.3, 12.4, 11.9, 12.4, 13.4),
cSEM = c(1.8, 1.8, 1.8, 1.8, 1.7, 1.7, 1.9, 1.7, 1.8, 2.1),
tAvgCPM = c(6.6, 6.6, 6.6, 6.6, 6.6, 7.4, 7.4, 7.1, 7.2, 8.0),
tSEM = c(0.7, 0.8, 0.8, 0.8, 0.8, 1.0, 1.0, 0.9, 0.5, 0.8)
)
I always want my Control group to be plotted in black and my Treatment group to be plotted in red. ConditionID can be used to identify the appropriate group for a dataset but it is not the value that I want displayed in the Legend.
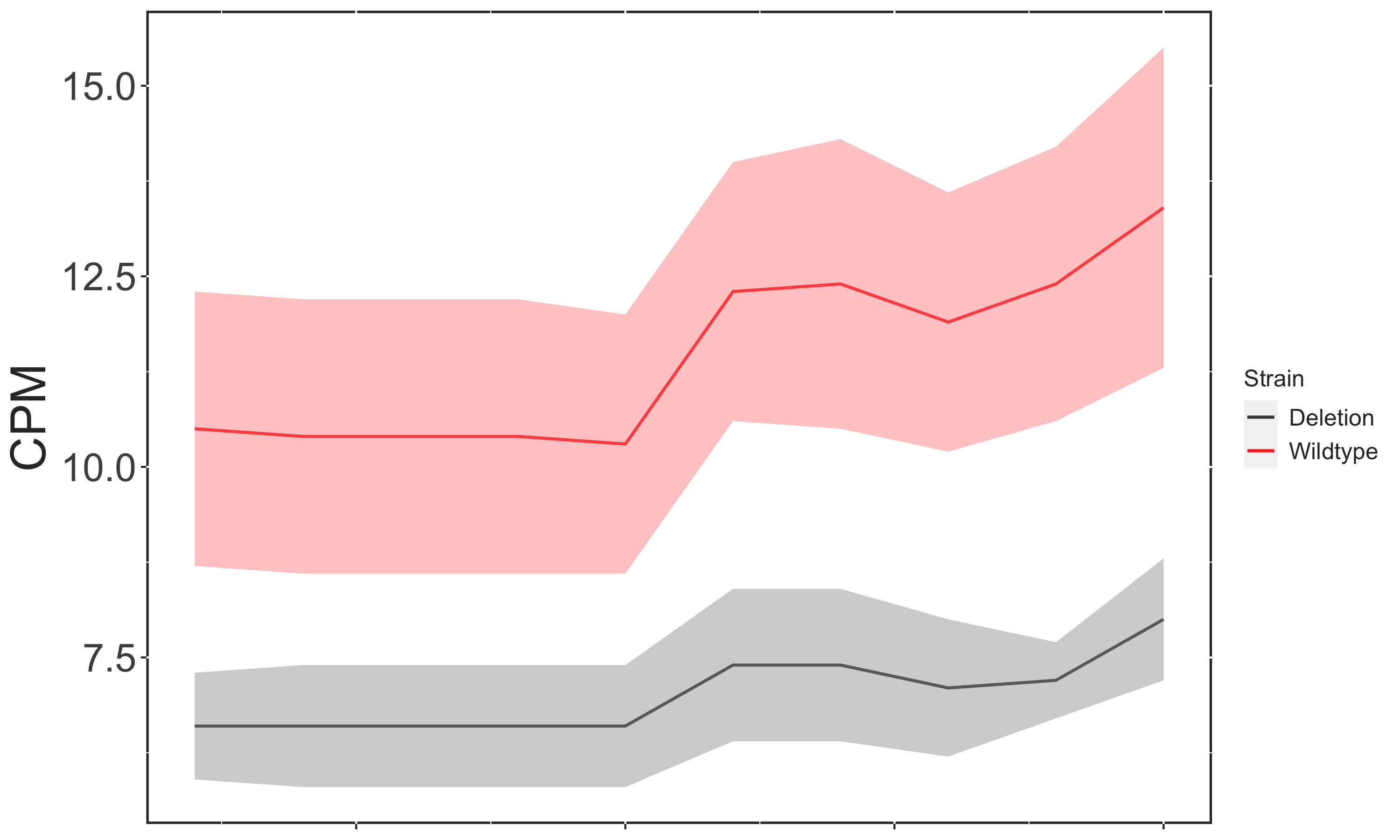
Here is an example that generates the plot exactly how I want it, but this is only true because the values in LUT$Treatment are in the correct alphabetical order.
Plot Correct
plot <- ggplot()
geom_ribbon(data = data, aes(x=bp, ymax=cAvgCPM cSEM, ymin=cAvgCPM-cSEM, fill = paste0(LUT$Treatment[LUT$ConditionID == "Control_Rep1"])), size = 1.2, alpha = 0.25)
geom_line(data = data, aes(x=bp, y=cAvgCPM, col = paste0(LUT$Treatment[LUT$ConditionID == "Control_Rep1"])), size = .75, alpha = 0.7)
geom_ribbon(data = data, aes(x=bp, ymax=tAvgCPM tSEM, ymin=tAvgCPM-tSEM, fill = paste0(LUT$Treatment[LUT$ConditionID == "Treatment_Rep1"])), size = 1.2, alpha = 0.25)
geom_line(data = data, aes(x=bp, y=tAvgCPM, col = paste0(LUT$Treatment[LUT$ConditionID == "Treatment_Rep1"])), size = .75, alpha = 0.7)
scale_fill_manual(values = c("#333333", "red"))
scale_color_manual(values = c("#333333", "red"))
ylab("CPM")
theme(text = element_text(family = "Arial", color = "#545454", size = 25),
panel.background = element_rect("white", color = "#333333", size = 1.2),
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.title.y = element_text(colour = "#333333"),
legend.title = element_text(colour = "#333333", size=12),
legend.text = element_text(colour = "#333333", size=12))
guides(fill = "none")
guides(color=guide_legend(title="Strain"))
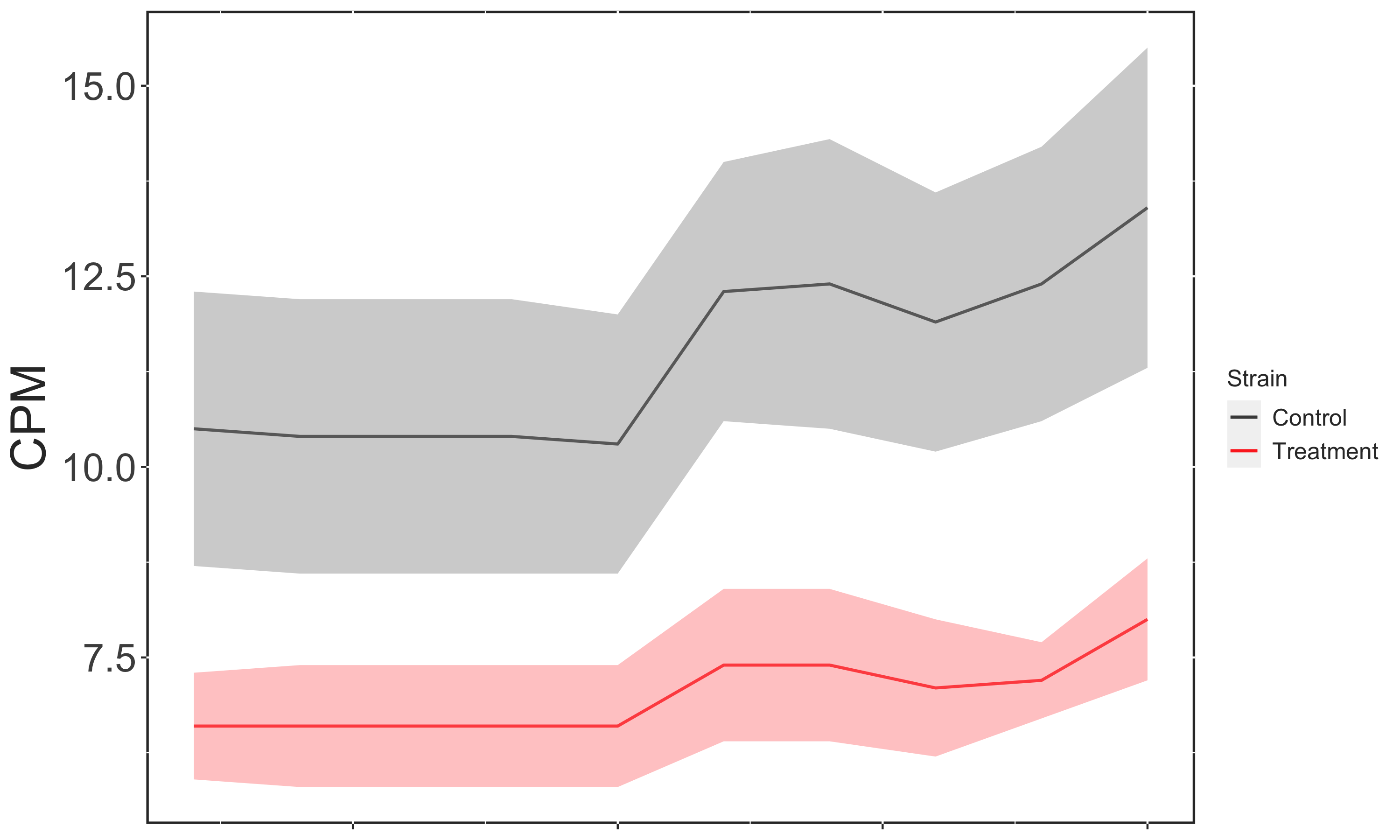 However, if I instead want to include LUT$Mutation in my Legend the plot now displays the incorrect coloring: Wildtype (the Control condition) is now red.
However, if I instead want to include LUT$Mutation in my Legend the plot now displays the incorrect coloring: Wildtype (the Control condition) is now red.
Plot Incorrect
plot <- ggplot()
geom_ribbon(data = data, aes(x=bp, ymax=cAvgCPM cSEM, ymin=cAvgCPM-cSEM, fill = paste0(LUT$Mutation[LUT$ConditionID == "Control_Rep1"])), size = 1.2, alpha = 0.25)
geom_line(data = data, aes(x=bp, y=cAvgCPM, col = paste0(LUT$Mutation[LUT$ConditionID == "Control_Rep1"])), size = .75, alpha = 0.7)
geom_ribbon(data = data, aes(x=bp, ymax=tAvgCPM tSEM, ymin=tAvgCPM-tSEM, fill = paste0(LUT$Mutation[LUT$ConditionID == "Treatment_Rep1"])), size = 1.2, alpha = 0.25)
geom_line(data = data, aes(x=bp, y=tAvgCPM, col = paste0(LUT$Mutation[LUT$ConditionID == "Treatment_Rep1"])), size = .75, alpha = 0.7)
scale_fill_manual(values = c("#333333", "red"))
scale_color_manual(values = c("#333333", "red"))
ylab("CPM")
theme(text = element_text(family = "Arial", color = "#545454", size = 25),
panel.background = element_rect("white", color = "#333333", size = 1.2),
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.title.y = element_text(colour = "#333333"),
legend.title = element_text(colour = "#333333", size=12),
legend.text = element_text(colour = "#333333", size=12))
guides(fill = "none")
guides(color=guide_legend(title="Strain"))
I've tried:
- factoring to add levels
- Adding variabled such as
scale_colour_discrete,scale_colour_identity - Assigning colors for each plot outside of
aes()
For instance, adding levels to LUT$Mutation still produces a plot with the Wildtype plotted in red.
LUT <- LUT %>%
dplyr::mutate(Mutation = factor(Mutation, levels=c("Wildtype", "Deletion")))
CodePudding user response:
You had some unneeded paste0's that I removed, I also refactored the fill/colour labelling variables so they can be set once and reused, and also used them to construct the colorscale.Finally I think its better to control the guides such that color is none and its fill that is renamed to strain
(from_lut <- LUT$Mutation[LUT$ConditionID == "Control_Rep1"])
(from_lut2 <- LUT$Mutation[LUT$ConditionID == "Treatment_Rep1"])
(colorscale <- c("#333333", "red"))
names(colorscale) <- c(from_lut,from_lut2)
colorscale
plot <- ggplot()
geom_ribbon(data = data, aes(x=bp, ymax=cAvgCPM cSEM, ymin=cAvgCPM-cSEM, fill = from_lut), size = 1.2, alpha = 0.25)
geom_line(data = data, aes(x=bp, y=cAvgCPM, col = from_lut), size = .75, alpha = 0.7)
geom_ribbon(data = data, aes(x=bp, ymax=tAvgCPM tSEM, ymin=tAvgCPM-tSEM, fill = from_lut2), size = 1.2, alpha = 0.25)
geom_line(data = data, aes(x=bp, y=tAvgCPM, col = from_lut2), size = .75, alpha = 0.7)
scale_fill_manual(values = colorscale)
scale_color_manual(values = colorscale)
ylab("CPM")
theme(text = element_text(family = "Arial", color = "#545454", size = 25),
panel.background = element_rect("white", color = "#333333", size = 1.2),
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.title.y = element_text(colour = "#333333"),
legend.title = element_text(colour = "#333333", size=12),
legend.text = element_text(colour = "#333333", size=12))
guides(fill=guide_legend(title="Strain")) guides(color = "none")