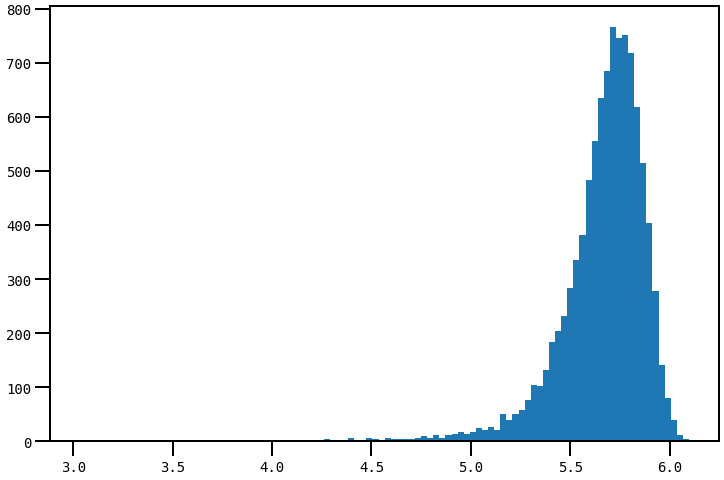
Consider
xdata=np.random.normal(5e5,2e5,int(1e4))
plt.hist(np.log10(xdata), bins=100)
plt.show()
plt.semilogy(xdata)
plt.show()
is there any way to display xticks of the first plot (plt.hist) as in the second plot's yticks? For good reasons I want to histogram the np.log10(xdata) of xdata but I'd like to set minor ticks to display as usual in a log scale (even considering that the exponent is linear...)
In other words, I want the x_axis of this plot:
to be like the y_axis
of the 2nd plot, without changing the spacing between major ticks (e.g., adding log marks between 5.5 and 6.0, without altering these values)
CodePudding user response:
yes you need to set the axes as log
ax = plt.gca() # get current axes
ax.set_yscale("log")
EDIT
Please note that plt module has not method set_yscale, if you don't want to recover the axes and alternative can be:
fig, ax = plt.subplots()
plt.hist(xdata) # equivalent to ax.hist(xdata)
ax.set_yscale("log")
plt.show()
CodePudding user response:
Just kept for now for clarification purpose. Will be deleted when the question is revised.
Disclaimer:
- As
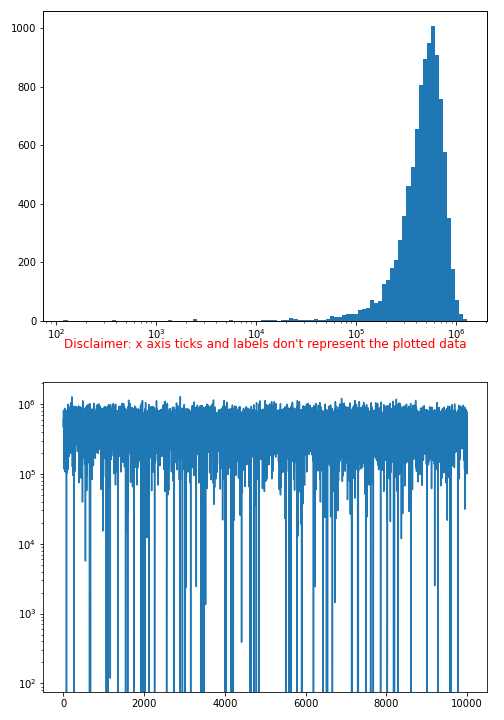
Explanation how that special axis transfer plot is done:
- original x-axis is hidden
- a
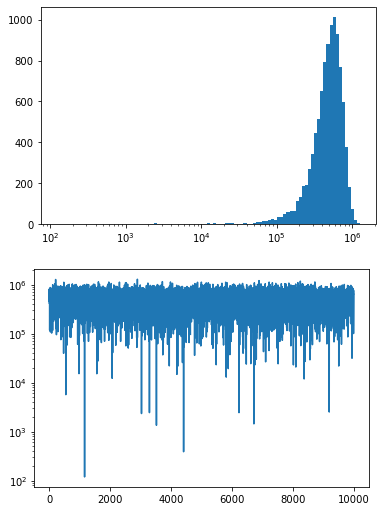
Explanation:
- Cut off negative values
- The randomly generated example data likely contains still some negative values
- activate the commented code lines at the beginning to see the effect
- logarithmic function isn't defined for values <= 0
- while the 2nd plot just deals with y-axis log scaling (negative values are just out of range), the 1st plot doesn't work with negative values in the BINs range
- probably real world working data won't be <= 0, otherwise keep that in mind
- The randomly generated example data likely contains still some negative values
- BINs should be aligned to log scale as well
- otherwise the 'BINs widths' distribution looks off
- switch
#on theplt.hist(statements in the 1st plot section to see the effect)
- switch
- otherwise the 'BINs widths' distribution looks off
xdata(notnp.log10(xdata)) to be plotted in the histogram- that 'workaround' with plotting
np.log10(xdata)probably was the root cause for the misunderstanding in the comments
- that 'workaround' with plotting
Code:
import numpy as np import matplotlib.pyplot as plt np.random.seed(42) # just to have repeatable results for the answer xdata=np.random.normal(5e5,2e5,int(1e4)) # MIN_xdata, MAX_xdata = np.min(xdata), np.max(xdata) # print(f"{MIN_xdata}, {MAX_xdata}") # note the negative values # cut off potential negative values (log function isn't defined for <= 0 ) xdata = np.ma.masked_less_equal(xdata, 0) MIN_xdata, MAX_xdata = np.min(xdata), np.max(xdata) # print(f"{MIN_xdata}, {MAX_xdata}") # align the bins to fit a log scale bins = 100 bins_log_aligned = np.logspace(np.log10(MIN_xdata), np.log10(MAX_xdata), bins) # 1st plot plt.hist(xdata, bins = bins_log_aligned) # note: xdata (not np.log10(xdata) ) # plt.hist(xdata, bins = 100) plt.xscale('log') plt.show() # 2nd plot plt.semilogy(xdata) plt.show() - Cut off negative values