I am trying to use R to make a line graph of time-series data for a genes expression level in two replicates (Rep2 and Rep3) over the course of 12-72 hours (points every two hours). I am a beginner at R and have looked at examples of making other time-series graphs on here and am still confused. I would appreciate any help you can give!
Here is what my dataframe looks like:

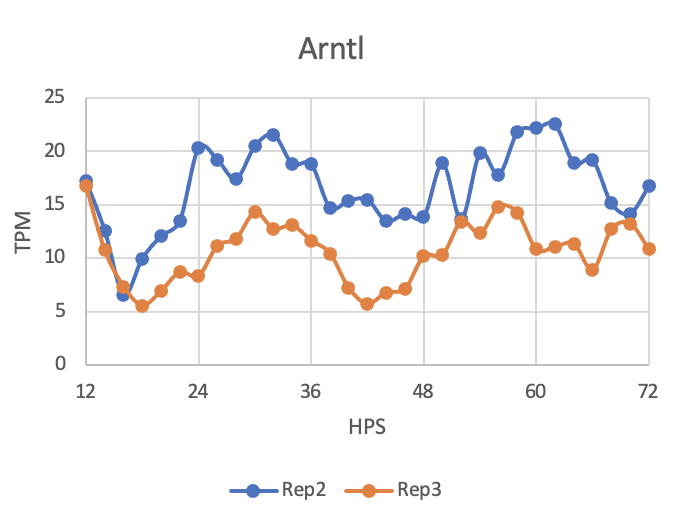
Here is what I'd like my final graph to look like:
CodePudding user response:
Without the data to test this, I can't know if it will work. I didn't feel like transcribing the photo of your spreadsheet. I feel like this will get you 90% of the way to the plot you show. The key is to go from wide format to long and then parse the numbers so they can be plotted.
library(tidyverse)
df |>
pivot_longer(cols = contains("X"), names_to = "HPS", values_to = "TPM")|>
mutate(HPS = parse_number(HPS))|>
ggplot(aes(HPS, TPM, color = gene_id))
geom_point()
geom_line()
ggtitle("Arntl")
scale_color_manual(values = c("blue", "orange"))
theme_bw()
theme(legend.position = "bottom",
legend.direction = "horizontal")
