I have a huge dataframe (2 million rows) in which a certain column has a string representations of a list of dictionaries (it is the school history of several people). So, what I'm trying to do is parsing this data to a new dataframe (because the relation is going to be 1 person to many schools).
However, my first option was to loop over the dataframe with itertuples(). Too slow!
The first few rows look like this:
list_of_dicts = {
0: '[]',
1: "[{'name': 'USA Health', 'subject': 'Residency, Internal Medicine, 2006 - 2009'}, {'name': 'Ross University School of Medicine', 'subject': 'Class of 2005'}]",
2: "[{'name': 'Physicians Medical Center Carraway', 'subject': 'Residency, Surgery, 1957 - 1960'}, {'name': 'Physicians Medical Center Carraway', 'subject': 'Internship, Transitional Year, 1954 - 1955'}, {'name': 'University of Alabama School of Medicine', 'subject': 'Class of 1954'}]"
}
df_dict = pd.DataFrame.from_dict(list_of_dicts, orient='index', columns=['school_history'])
What I thought about, was to have a function and then apply it to the dataframe:
def parse_item(row):
eval_dict = eval(row)[0]
school_df = pd.DataFrame.from_dict(eval_dict, orient='index').T
return school_df
df['column'].apply(lambda x: parse_item(x))
However, I'm not able to figure out how to generate a dataframe bigger than original (due to situations of multiple schools to one person).
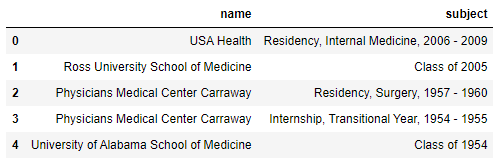
From those 3 rows, the idea is to have this dataframe (that has 5 rows from 2 rows):
CodePudding user response:
Iterate over the column to convert each string into a python list using ast.literal_eval(); the result is a nested list, which can be flattened inside the same comprehension.
N.B. Converting the column to a list (via tolist()) first sees some performance gain.
from ast import literal_eval
result = pd.DataFrame([x
for row in df_dict['school_history'].tolist()
for x in literal_eval(row)])
To retain the original index, instead of iterating over the column as a list, iterate over the zip object created by calling items() method on it. This returns (index, value) tuples where the indexes may be attached to the final output values.
ind, data = zip(*[(i, x)
for i, row in df_dict['school_history'].items()
for x in ast.literal_eval(row)]);
result = pd.DataFrame(data, index=ind)
CodePudding user response:
This does the trick using your sample data (thanks for the performance tip in comments):
list_df = df_dict.school_history.map(ast.literal_eval)
exploded = list_df[list_df.str.len() > 0].explode()
final = pd.DataFrame(list(exploded), index=exploded.index)
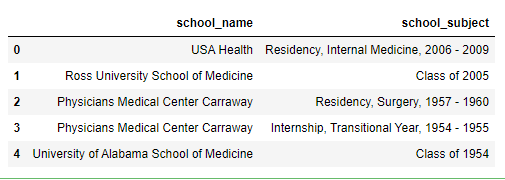
This produces the following:
In [54]: final
Out[54]:
name subject
1 USA Health Residency, Internal Medicine, 2006 - 2009
1 Ross University School of Medicine Class of 2005
2 Physicians Medical Center Carraway Residency, Surgery, 1957 - 1960
2 Physicians Medical Center Carraway Internship, Transitional Year, 1954 - 1955
2 University of Alabama School of Medicine Class of 1954
This will probably not be super fast given the amount of data, but parsing a dictionary of strings with nested objects inside will probably be pretty slow no matter what. You're probably better off parsing the file upstream first, then converting to pandas.