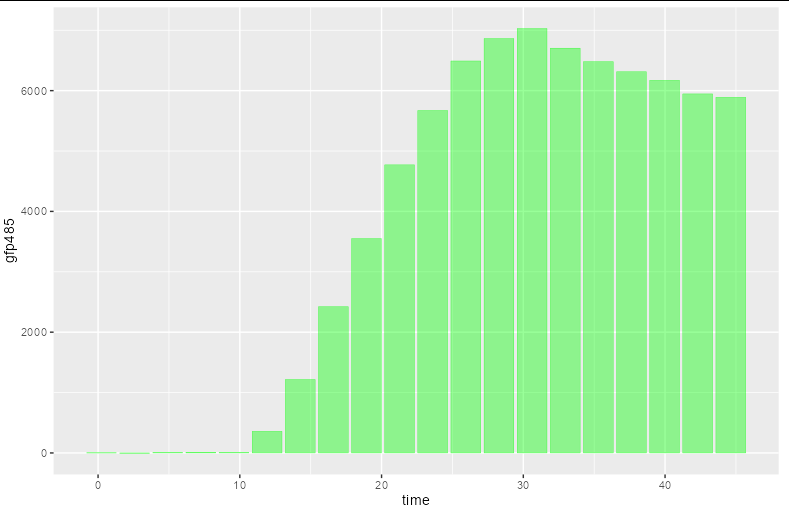
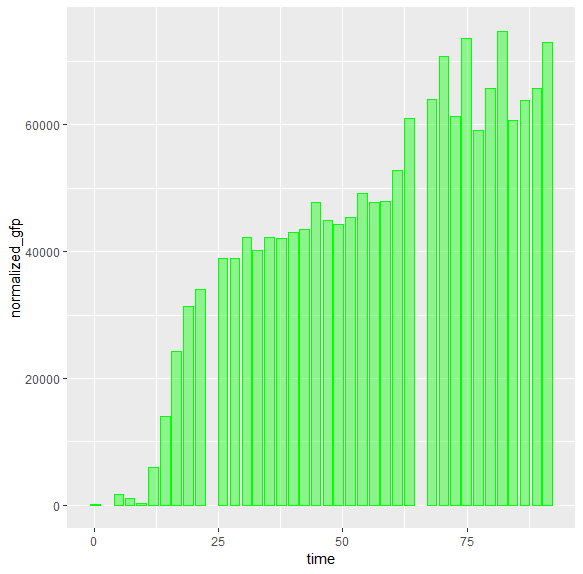
I'm currently in the process of making bar charts from experimental data. It seems that when I plot the bars on a continuous x-scale the bars will have uneven spacing/gaps between them. I would assume this is because of missing data points for specific times, but is there any way to make the gaps even?
I used the following simple code to plot this:
dataset_test %>%
ggplot(aes(time, normalized_gfp))
geom_bar(stat="identity", size=.1, fill="green", color="green", alpha=.4)
Example of gfp485 bar chart plot:
[ ]
]
Head of data sample (I'm plotting gfp485 in this example, but I'm gonna plot "od" on top in the final plot)
> head(dataset,20)
time media gfp485 od
1 0.24 IO CasA_gfp 0.3333333 0.006666667
2 2.64 IO CasA_gfp -4.3333333 0.003333333
3 5.04 IO CasA_gfp 5.6666667 0.003333333
4 7.20 IO CasA_gfp 10.6666667 0.010000000
5 9.60 IO CasA_gfp 6.3333333 0.023333333
6 12.00 IO CasA_gfp 358.3333333 0.060000000
7 14.40 IO CasA_gfp 1216.6666667 0.086666667
8 16.56 IO CasA_gfp 2422.6666667 0.100000000
9 18.96 IO CasA_gfp 3550.3333333 0.113333333
10 21.36 IO CasA_gfp 4770.3333333 0.140000000
11 23.52 IO CasA_gfp 5671.3333333 NA
12 25.92 IO CasA_gfp 6491.0000000 0.166666667
13 28.32 IO CasA_gfp 6862.6666667 0.176666667
14 30.72 IO CasA_gfp 7028.3333333 0.166666667
15 32.88 IO CasA_gfp 6704.0000000 0.166666667
16 35.28 IO CasA_gfp 6480.3333333 0.153333333
17 37.68 IO CasA_gfp 6312.6666667 0.150000000
18 40.08 IO CasA_gfp 6171.0000000 0.143333333
19 42.24 IO CasA_gfp 5945.3333333 0.136666667
20 44.64 IO CasA_gfp 5889.6666667 0.123333333
Thank you very much in advance :))
CodePudding user response:
We can see the uneven spacing of your bars in the sample data, even without missing values:
library(ggplot2)
ggplot(dataset_test, aes(time, gfp485))
geom_col(size= .1, fill = "green", color = "green", alpha = 0.4)
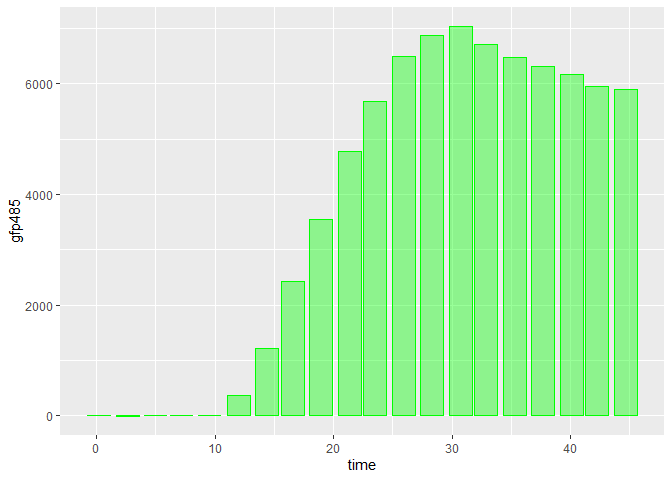
The reason for this is that your observations are not evenly spaced in time. If we check the difference between consecutive time values, we will see they are not all the same:
diff(dataset_test$time)
#> [1] 2.40 2.40 2.16 2.40 2.40 2.40 2.16 2.40 2.40 2.16 2.40 2.40 2.40 2.16
#> [15] 2.40 2.40 2.40 2.16 2.40
If you are prepared to change the actual data for a prettier plot, but keep the overall time equal to the original, you could do:
ggplot(dataset_test,
aes(x = min(time) seq(0, by = mean(diff(time)), length = length(time)),
y = gfp485))
geom_col(size= .1, fill = "green", color = "green", alpha = 0.4)
labs(x = "time")
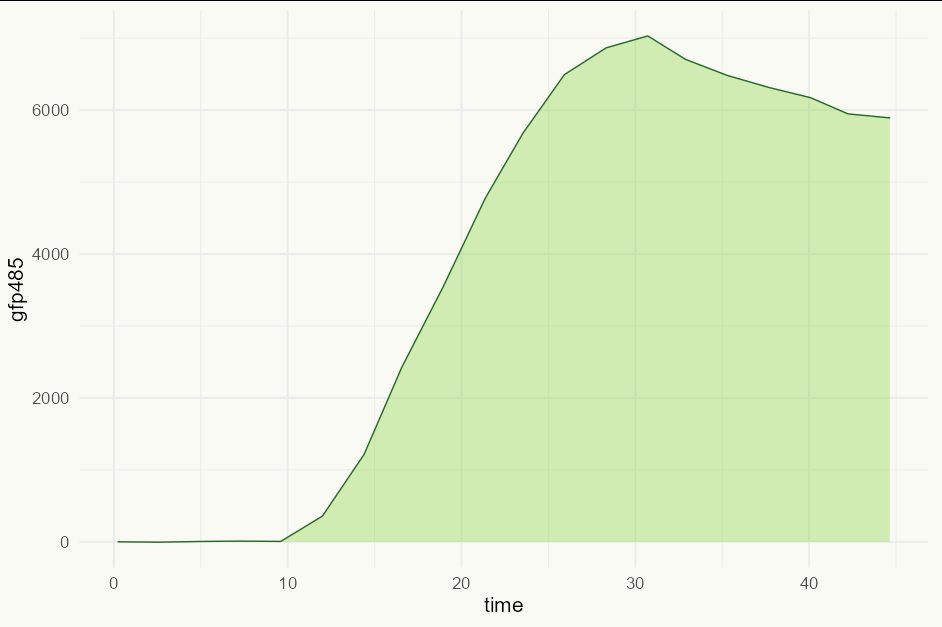
However, if you have unequally spaced time data and a continuous variable on the y axis, then it would be more honest (and, I would argue, more visually appealing) to use geom_area:
ggplot(dataset_test, aes(time, gfp485))
geom_area(fill = "#90d850", color = "#266825", alpha = 0.4, size = 0.5)
theme_minimal(base_size = 16)
theme(plot.background = element_rect(fill = "#fafaf4", color = NA))
Created on 2022-10-11 with reprex v2.0.2