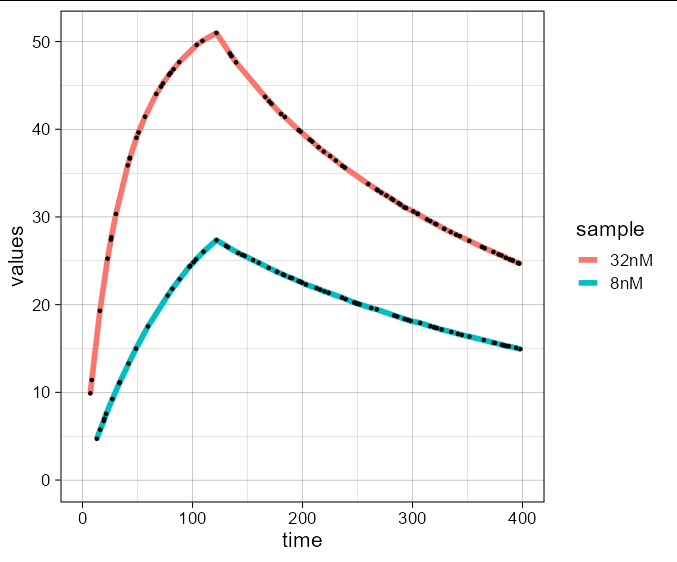
I am trying to fit association-dissociation SPR kinetics data for a protein and small molecule for two concentrations using ggplot2. The data is 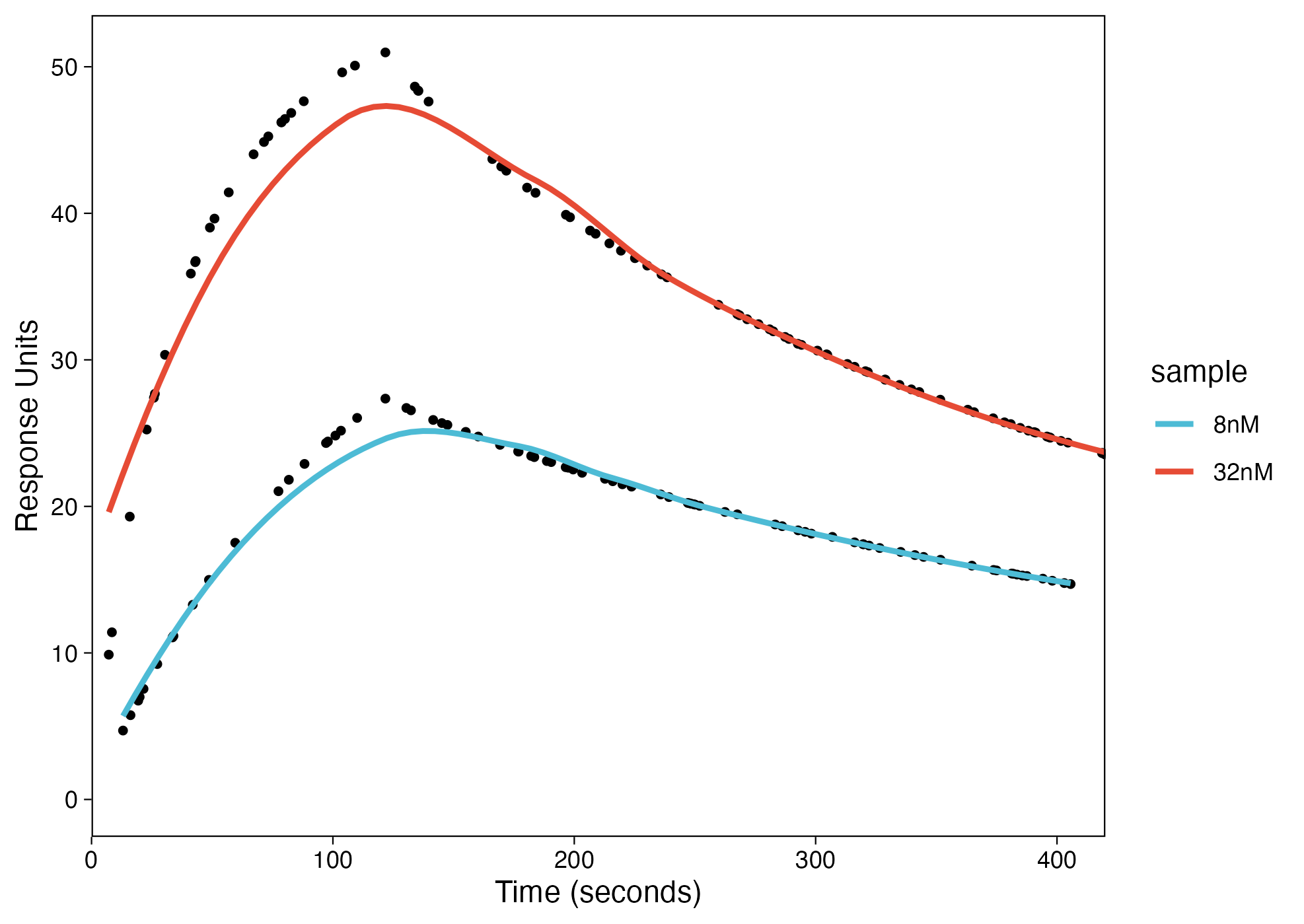
As you can see that the fit did not work using method = "loess". I need something like this(there are 5 concentrations here):
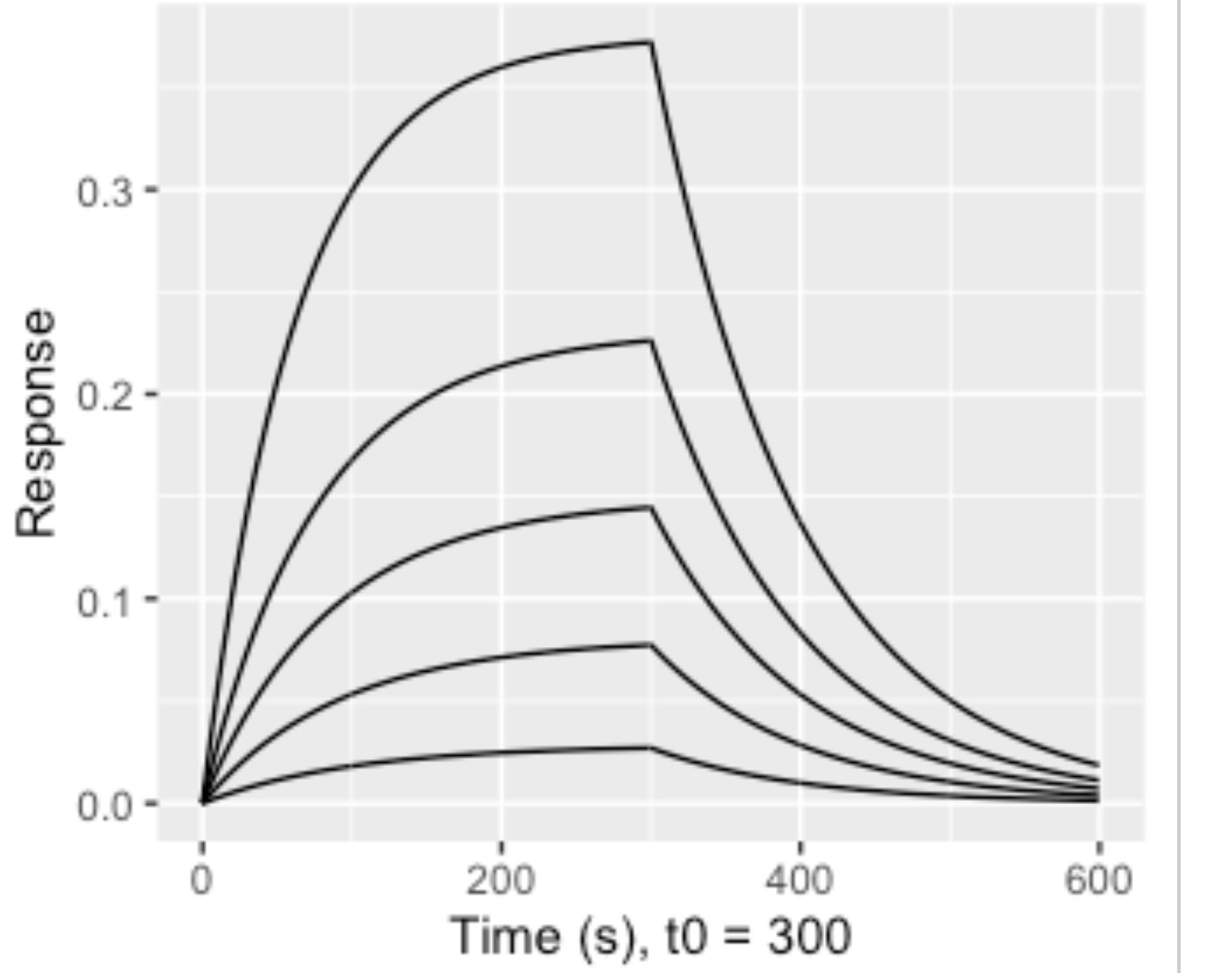
The fitting requires 1:1 Langmuir model but I am not sure how I can do that in ggplot. Can someone please help me?
Edit
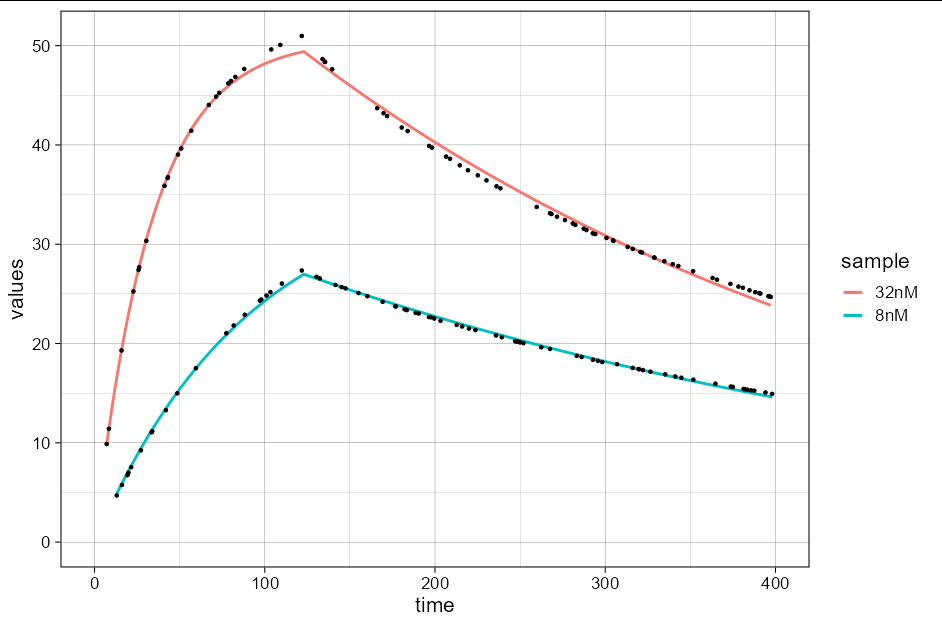
It is possible to fit the results to the data using non-linear least squares, employing the binding1to1 function from pbm, but it requires a bit of method tweaking to get the model to fit. It would probably be better to create a model then plot the predictions rather than using geom_smooth. However, if you really wanted to, you could do:
df %>%
ggplot(aes(time, values, color = sample))
geom_smooth(method = nls, se = FALSE, n = 1000,
formula = y ~ binding1to1(x, 123, 32e-9, kon, koff, rmax),
method.args = list(
start = list(kon = 2000, koff = 0.02, rmax = 2e4),
control = nls.control(minFactor = 1e-6, maxiter = 1000)
),
data = df[df$time > 0 & df$sample == "32nM",])
geom_smooth(method = nls, se = FALSE, n = 1000,
formula = y ~ binding1to1(x, 123, 8e-9, kon, koff, rmax),
method.args = list(
start = list(kon = 3000, koff = 0.02, rmax = 2e4),
control = nls.control(minFactor = 1e-9, maxiter = 10000)
),
data = df[df$time > 0 & df$sample == "8nM",])
geom_point(color = 'black', size = 1)
theme_linedraw(base_size = 16)
xlim(c(0, 400))
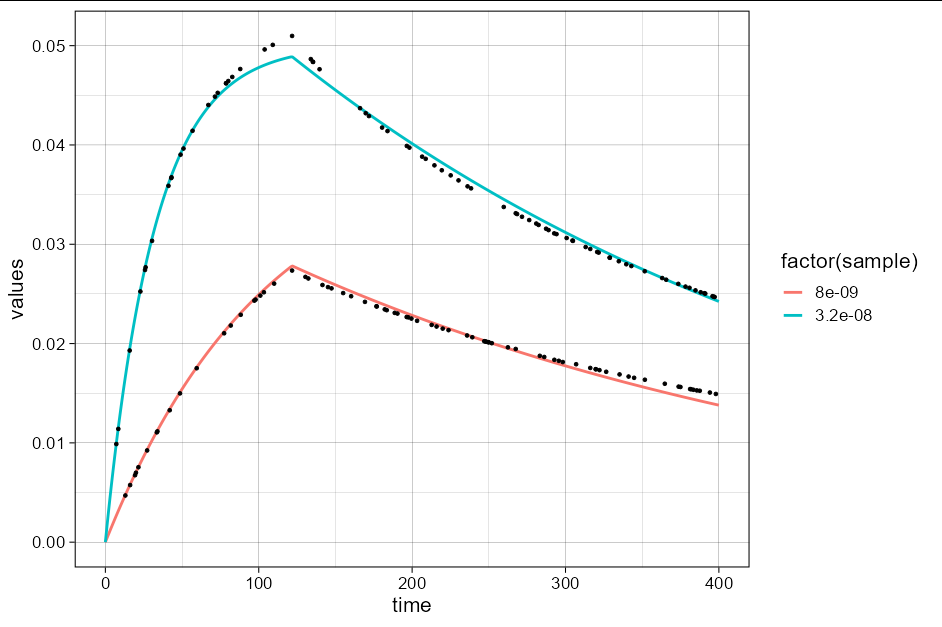
If you want to actually fit a model from which to extract the parameters and plot, you can do:
library(tidyverse)
library(pbm)
df <- read.csv("SPR.csv") %>%
filter(time >= 0) %>%
mutate(sample = as.numeric(gsub("\\D ", "", sample)) * 1e-9,
values = values * 1e-3) %>%
group_by(sample) %>%
mutate(tmax = time[which.max(values)])
fit_fun <- function(time, tmax, sample, kon, koff, rmax) {
unlist(Map(function(time, tmax, sample) {
binding1to1(time, tmax, sample, kon, koff, rmax)
}, time, tmax, sample))
}
mod <- nls(values ~ fit_fun(time, tmax, sample, kon, koff, rmax),
data = df,
start = list(kon = 3000, koff = 0.02, rmax = 2),
control = nls.control(minFactor = 1e-9, maxiter = 10000))
This gives us a model with the best fitting values for the various parameters:
mod
#> Nonlinear regression model
#> model: values ~ fit_fun(time, tmax, sample, kon, koff, rmax)
#> data: df
#> kon koff rmax
#> 8.925e 05 2.521e-03 5.445e-02
#> residual sum-of-squares: 5.219e-05
#>
#> Number of iterations to convergence: 536
#> Achieved convergence tolerance: 5.155e-07
We can then predict the output of the model over the range of our input variables:
pred_df <- expand.grid(time = 0:400, sample = c(8, 32) * 1e-9,
tmax = df$tmax[1])
pred_df$values <- predict(mod, pred_df)
And we can plot it like this:
df %>%
ggplot(aes(time, values, color = factor(sample)))
geom_line(data = pred_df, size = 1)
geom_point(color = 'black', size = 1)
theme_linedraw(base_size = 16)
xlim(c(0, 400))