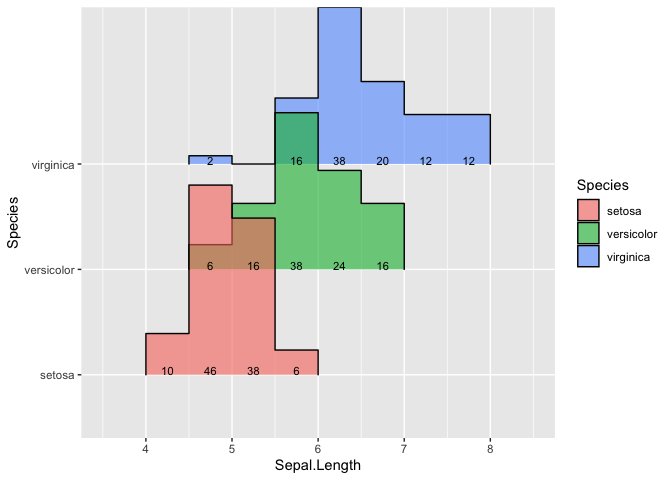
I would like to use ggridges to plot a binned ridgeline, with the percentage of each bin labelled to the bins. I have attempted to use geom_text(stat ="bin) to calculate percentages, but the calculation use all the data. I would like to calculate the percentage separately for each species. Below is the code and the output.
iris_mod=rbind(iris, iris[iris$Species=="setosa",])
#This adds more setosa, so the distribution becomes 100,50, and 50.
ggplot(iris_mod,aes(x=Sepal.Length, y=Species, fill=Species))
geom_density_ridges(alpha=0.6, stat="binline", binwidth = .5, draw_baseline = FALSE,boundary = 0)
geom_text(
stat = "bin",
aes(y = group 0*stat(count/count),
label = round(stat(count/sum(count)*100),2)),
vjust = 0, size = 3, color = "black", binwidth = .5, boundary=0)
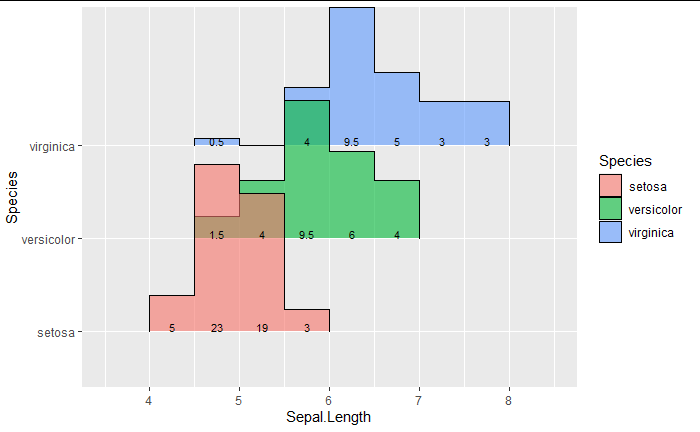
As you can see from the setosa labels, its 5, 23, 19, 3 which adds up to 50, while the other two adds up to 25 each. I wanted the setosa labels to be 10, 46, 38 and 6, which should add up to 100, and the other two species to add up to 100 as well.
CodePudding user response:
Using e.g. tapply to compute sum per group and a small custom function you could do:
library(ggplot2)
library(ggridges)
iris_mod <- rbind(iris, iris[iris$Species == "setosa", ])
comp_pct <- function(count, group) {
label <- count / tapply(count, group, sum)[as.character(group)] * 100
ifelse(label > 0, round(label, 2), "")
}
ggplot(iris_mod, aes(x = Sepal.Length, y = Species, fill = Species))
geom_density_ridges(alpha = 0.6, stat = "binline", binwidth = .5, draw_baseline = FALSE, boundary = 0)
geom_text(
stat = "bin",
aes(
y = after_stat(group),
label = after_stat(comp_pct(count, group))
),
vjust = 0, size = 3, color = "black", binwidth = .5, boundary = 0
)