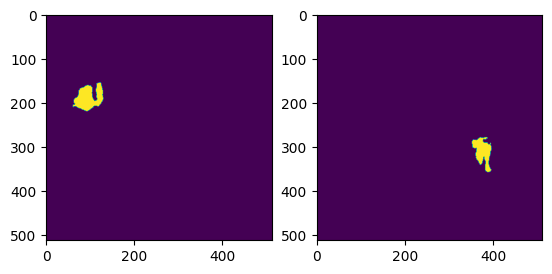
I have a numpy array with dimensions (512, 512, 3), which represents an RGB image. The image is a simple black and white mask, visualisation:

My goal is to identify the x largest clusters of white pixels in this mask and produce new images exclusive to each cluster. For example, for the image above, if x = 2, my desired algorithm would produce the following 2 images:

Is there a way I can do this simply with numpy? My naive approach has been to use depth-first-search.
CodePudding user response:
find your points like this:
im = Image.open('Nkuf9.png')
data = np.array(im)[:,:,0]
wpoint = np.where(data == 255)
points = set((x, y) for x, y in zip(*wpoint) )
and a simple dfs to find each connected region, you could put all in a class.
def generate_neighbours(point):
neighbours = [
(1, -1), (1, 0),(1, 1),
(0, -1), (0, 1),
(1, -1), (1, 0),(-1, 1)
]
for neigh in neighbours:
yield tuple(map(sum, zip(point, neigh)))
def find_regions(p , points):
reg = []
seen = set()
def dfs(point):
if point not in seen:
seen.add(point)
if point in points:
reg.append(point)
points.remove(point)
for n in generate_neighbours(point):
dfs(n)
dfs(p)
return reg
region =[]
while points:
cur = next(iter(points))
reg = find_regions(cur, points)
region.append(reg.copy())
areas = {idx: area for idx, area in enumerate(map(len,region))}
areas = sorted(areas.items(), key=lambda x: x[1], reverse=True)
num = 2
for idx, area in enumerate(areas[:num]):
plt.subplot(1,num, idx 1)
im = np.zeros((512, 512))
for x,y in region[area[0]]:
im[x,y] = 255
plt.imshow(im)