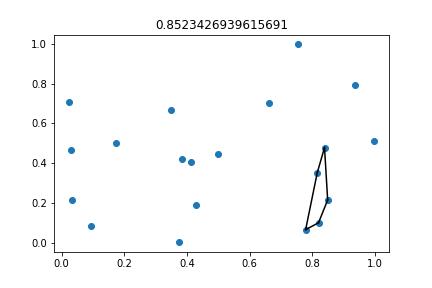
I have many (x, y) points. I want to fit the smallest (or an approximation or estimation of the smallest) convex hull around a variable percentage of those points.
The existing implementations I can find get the convex hull around all the points. How can I have a function like so:
def get_convec_hull(points, percentage):
# some code here
return smallest_hull
I can brute force my desired outcome using the code below. This does exactly what I want, except that it's really slow. Finding the smallest convex hull around a portion of 25% of 40 data points already takes a couple of sections. My real data has many million points.
# A brute force solution..
from scipy.spatial import ConvexHull
import itertools
import numpy as np
import math
np.random.seed(11111)
# Simulating my data..
n_points = 20
target_percentage = 0.25
points = np.random.random((n_points, 2))
def get_convec_hull(points, percentage):
n_points = points.shape[0]
target_points = int(n_points * percentage)
print("Looking for smallest polygon covering {} of the {} datapoints!".format(target_points, n_points))
print(target_points)
subsets = itertools.combinations(list(range(n_points)), target_points)
optimal = ConvexHull(points[next(subsets),:]) # first one designated as optimal at the start
for subset in subsets:
hull = ConvexHull(points[subset,:])
if hull.area < optimal.area:
optimal = hull
return optimal
optimal = get_convec_hull(points, target_percentage)
optimal.area # returns 0.85234...
Also, just to illustrate, this is the 'optimal', i.e., the smallest convex hull around 25% (5 points) for the simulated dataset.
How could I apply this to a much larger dataset?
CodePudding user response:
Please realize I am risking my life posting this; I am fairly certain actual computer scientists are already tracing my IP.
I have rewritten your function in the following manner:
def get_suboptimal_hull(points,p,magic_number):
n_points = points.shape[0]
tp = int(n_points * p)
d_mat = distance_matrix(points,points)
d_mat_s = d_mat.copy()
d_mat_s.sort(axis=1)
dist = np.sum(d_mat_s[:,0:5],axis=1)
dens = d_mat_s[:,4]/(dist**2)
densstr = [str(x)[0:4] for x in dens]
dens_s = dens.copy()
dens_s.sort()
mnp = -1 * int(magic_number*points.shape[0]) - 1
suboptimal = points[dens > dens_s[mnp]]
n_points = len(suboptimal)
subsets = itertools.combinations(list(range(n_points)), tp)
optimal = ConvexHull(suboptimal[next(subsets),:])
for subset in subsets:
hull = ConvexHull(suboptimal[subset,:])
if hull.area < optimal.area:
optimal = hull
return optimal
The way I achieve the speedup is by "cleverly" pruning the point tree based on a density parameter. By pruning the tree with a divisor of magic_number, you can achieve a magic_number^2 divisor speedup (tree generation is N^2). There are two things to take care of:
- I still use your function to approach the optimal value
- The speedup and performance is heavily dependent on the value of
magic_number.
Currently, magic_number is a percentage of the points, but could just as easily be modified as a fixed number to stabilize performance. Might be useful for your applications.
Init:
from scipy.spatial import ConvexHull
from scipy.spatial import distance_matrix
import itertools
import numpy as np
import math
import matplotlib.pyplot as plt
np.random.seed(11111)
n_points = 20
target_percentage = 0.25
points = np.random.random((n_points, 2))
magic_number = 0.5
The area is the same (there could be exceptions where is is not true, methinks):
optimal = get_convec_hull(points, target_percentage)
suboptimal = get_suboptimal_hull(points, target_percentage,0.5)
print((optimal.area, suboptimal.area))
Returns:
(0.8523426939615691, 0.8523426939615691)
(I noticed that in this configuration, this is perimeter. Still the same tough.)
Speedup:
yours = %timeit -r 3 -o get_convec_hull(points, target_percentage)
mine = %timeit -r 3 -o get_suboptimal_hull(points, target_percentage,magic_number)
your_t = np.mean(yours.all_runs)
my_t = np.mean(mine.all_runs)
print(f'For magic number {magic_number}, expected {int(100*(1/magic_number**2))} % speedup, got a {int(100* (your_t/my_t))} % speedup.')
Returns:
8.44 s ± 162 ms per loop (mean ± std. dev. of 3 runs, 1 loop each)
143 ms ± 1.67 ms per loop (mean ± std. dev. of 3 runs, 10 loops each)
For magic number 0.5, expected 400 % speedup, got a 590 % speedup.