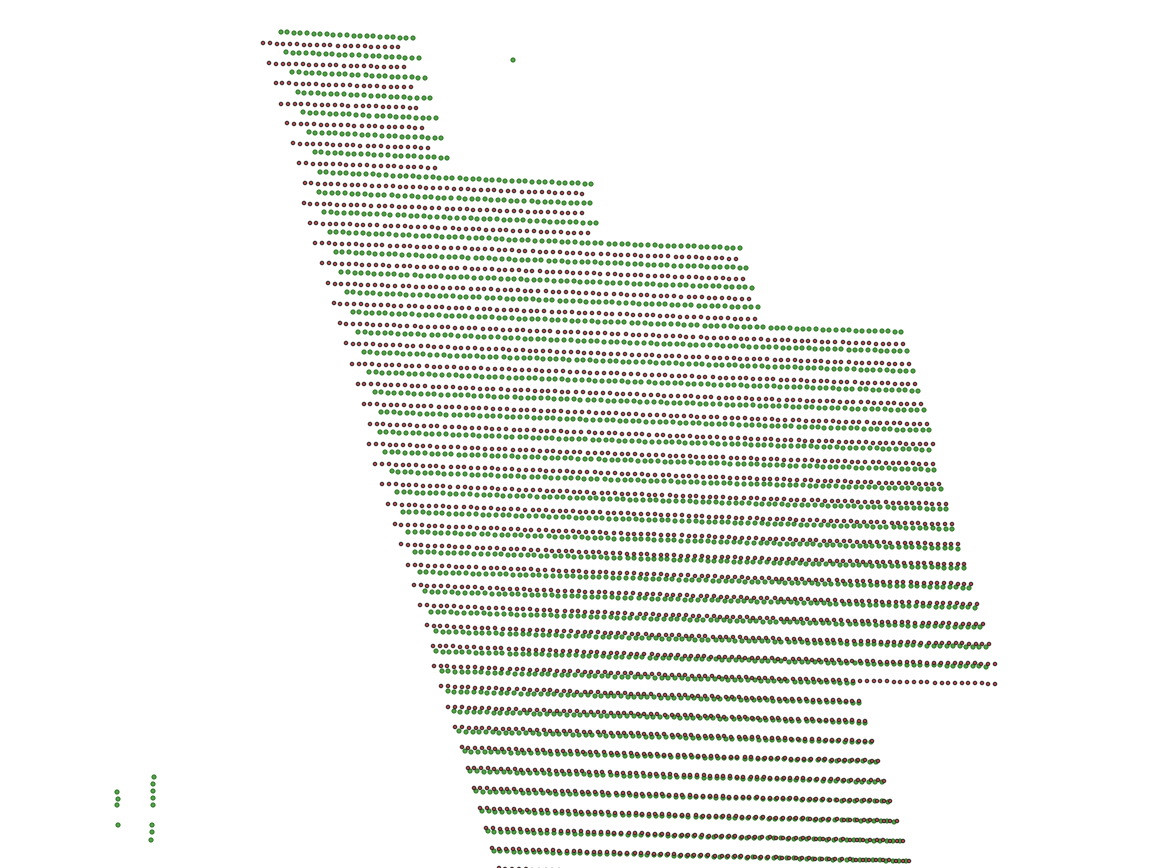
I have two large sets of (x, y) points and I want to associate in Python each point of one set with "the corresponding point" of the other.
The second set can also contain outliers, i.e. extra noise points, as you can see in this picture, where there are more green dots than red dots:
The association between the two sets of points is not a simple translation, as you can see in this image:
In these two links you can find the red dots and green dots (list of image coordinates with origin in top-left):
https://drive.google.com/file/d/1fptkxEDYbIJ93r_OXJSstDHMfk67DDYo/view?usp=share_link https://drive.google.com/file/d/1Z_ghWIzUZv8sxfawOBoGG3fJz4h_z7Qv/view?usp=share_link
My problem is similar to these two:
How to align two sets of points (translation rotation) when those sets contain noise?
but I have a large set of points, so the solutions proposed here don't work for my case. My points have a certain structure in rows so it's difficult to compute a Roto-Scale-Translation function because the rows of points get confused with each other.
CodePudding user response:
I found a method which can recover which points correspond to which other points fairly accurately, using two phases. The first phase corrects for affine transformation, and the second phase corrects for nonlinear distortion.
Note: I chose to match red points to green points, rather than the other way around.
Assumptions
The method makes three assumptions:
- It knows three or more green points and the matching red points.
- The differences between the two are mostly linear.
- The nonlinear portion of the difference is locally similar - i.e. if one point has the match offset by (-10, 10), the neighboring point will have a similar offset. This is controlled by
max_search_dist.
Code
Start by loading both datasets:
import json
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import numpy as np
from sklearn.neighbors import NearestNeighbors
from scipy.spatial import KDTree
from collections import Counter
with open('red_points.json', 'rb') as f:
red_points = json.load(f)
red_points = pd.DataFrame(red_points, columns=list('xy'))
with open('green_points.json', 'rb') as f:
green_points = json.load(f)
green_points = pd.DataFrame(green_points, columns=list('xy'))
I found it useful to have a function to visualize both datasets:
def plot_two(green, red):
if isinstance(red, np.ndarray):
red = pd.DataFrame(red, columns=list('xy'))
if isinstance(green, np.ndarray):
green = pd.DataFrame(green, columns=list('xy'))
both = pd.concat([green.assign(hue='green'), red.assign(hue='red')])
ax = both.plot.scatter('x', 'y', c='hue', alpha=0.5, s=0.5)
ax.ticklabel_format(useOffset=False)
Next, pick three points in green, and provide their XY coordinates. Find the corresponding points in red, and provide their XY coordinates.
green_sample = np.array([
[5221, 12460],
[2479, 2497],
[6709, 6303],
])
red_sample = np.array([
[5274, 12597],
[2375, 2563],
[6766, 6406],
])
Next, use those points to find an affine matrix. This affine matrix will cover rotation, translation, scaling and skewing. Since it has six unknowns, you need at least six constraints, or the equation is underdetermined. This is why we needed at least three points earlier.
def add_implicit_ones(matrix):
b = np.ones((matrix.shape[0], 1))
return np.concatenate((matrix,b), axis=1)
def transform_points_affine(points, matrix):
return add_implicit_ones(points) @ matrix
def fit_affine_matrix(red_sample, green_sample):
red_sample = add_implicit_ones(red_sample)
X, _, _, _ = np.linalg.lstsq(red_sample, green_sample, rcond=None)
return X
X = fit_affine_matrix(red_sample, green_sample)
red_points_transformed = transform_points_affine(red_points.values, X)
Now we get to the nonlinear matching step. This is run after red's values are transformed to match green's values. Here's the algorithm:
- Start at a red point which has no nonlinear component. One near one of the
green_samplepoints is a good choice for this - the affine step will prioritize getting those points right. Search around this point in a radius for the corresponding green point. Record the difference between the red point and the corresponding green point as "drift." - Look at all of the red neighbors of that red point. Add those to a list to process.
- At one of those red neighbors, take the average drift of all neighboring red points. Add that drift to the red point, and search in a radius for a green point.
- The difference between the red point and corresponding green point is the drift for this red point.
- Add all this point's red neighbors to the list to process, and go back to step 3.
def find_nn_graph(red_points_np):
nbrs = NearestNeighbors(n_neighbors=8, algorithm='ball_tree').fit(red_points_np)
_, indicies = nbrs.kneighbors(red_points_np)
return indicies
def point_search(red_points_np, green_points_np, starting_point, max_search_radius):
starting_point_idx = (((red_points_np - starting_point)**2).mean(axis=1)).argmin()
green_tree = KDTree(green_points_np)
dirty = Counter()
visited = set()
indicies = find_nn_graph(red_points_np)
# Mark starting point as dirty
dirty[starting_point_idx] = 1
match = {}
drift = np.zeros(red_points_np.shape)
# NaN = unknown drift
drift[:] = np.nan
while len(dirty) > 0:
point_idx, num_neighbors = dirty.most_common(1)[0]
neighbors = indicies[point_idx]
if point_idx != starting_point_idx:
neighbor_drift_all = drift[neighbors]
if np.isnan(neighbor_drift_all).all():
# All neighbors have no drift
# Unmark as dirty and come back to this one
del dirty[point_idx]
continue
neighbor_drift = np.nanmean(neighbor_drift_all, axis=0)
assert not np.isnan(neighbor_drift).any(), "No neighbor drift found"
else:
neighbor_drift = np.array([0, 0])
# Find the point in the green set
red_point = red_points_np[point_idx]
green_points_idx = green_tree.query_ball_point(red_point neighbor_drift, r=max_search_radius)
assert len(green_points_idx) != 0, f"No green point found near {red_point}"
assert len(green_points_idx) == 1, f"Too many green points found near {red_point}"
green_point = green_points_np[green_points_idx[0]]
real_drift = green_point - red_point
match[point_idx] = green_points_idx[0]
# Save drift
drift[point_idx] = real_drift
# Mark unvisited neighbors as dirty
if point_idx not in visited:
neighbors = indicies[point_idx, 1:]
neighbors = [n for n in neighbors if n not in visited]
dirty.update(neighbors)
# Remove this point from dirty
del dirty[point_idx]
# Mark this point as visited
visited.add(point_idx)
# Check that there are no duplicates
assert len(set(match.values())) == len(match)
# Check that every point in red_points_np was matched
assert len(match) == red_points_np.shape[0]
return match, drift
# This point is assumed to have a drift of zero
# Pick one of the points which was used for the linear correction
starting_point = green_sample[0]
# Maximum distance that a point can be found from where it is expected
max_search_radius = 10
green_points_np = green_points.values
match, drift = point_search(red_points_transformed, green_points_np, starting_point, max_search_radius)
Next, here's a tool which you can use to audit the quality of the match. This is showing the first thousand matches. Underneath that is a quiver plot, where the arrow points from the red point in the direction of the matching green point. (Note: arrows are not to scale.)
red_idx, green_idx = zip(*match.items())
def show_match_subset(start_idx, length):
end_idx = start_idx length
plot_two(green_points_np[np.array(green_idx)][start_idx:end_idx], red_points_np[np.array(red_idx)][start_idx:end_idx])
plt.show()
red_xy = red_points_np[np.array(red_idx)][start_idx:end_idx]
red_drift_direction = drift[np.array(red_idx)][start_idx:end_idx]
plt.quiver(red_xy[:, 0], red_xy[:, 1], red_drift_direction[:, 0], red_drift_direction[:, 1])
show_subset(0, 1000)
Plots:
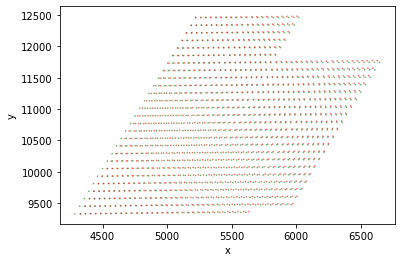
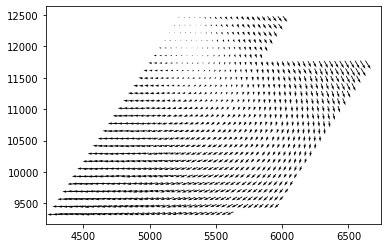
Match
Here's a copy of the match I found. It's in JSON format, where the keys represent indexes of points in the red point file, and the values represent indexes of points in the green point file. https://pastebin.com/SBezpstu