When I call the user defined function sRGB_to_CAM16UCS in the console, it displays the result as intended. But when I try to call it within a while loop it throws an error. Can somebody help me understand the error?
library(purrr)
library(tibble)
library(tidyr)
sRGB_to_CAM16UCS <- function(R255, G255, B255){
# Convert sRGB to 1931 CIE XYZ [IEC 61966-2–1:2003(E)]
## Convert to the range of 0 to 1
R1 <- R255 / 255
G1 <- G255 / 255
B1 <- B255 / 255
## Gamma Expansion of sRGB values
gamma_inverse <- function(RGB1){
if (RGB1 < -0.04045 | RGB1 > 0.04045){
((RGB1 0.055)/1.055)^2.4
} else {
RGB1/12.92
}
}
R_li <- gamma_inverse(R1)
G_li <- gamma_inverse(G1)
B_li <- gamma_inverse(B1)
# Convert linear RGB values to CIE XYZ
X <- 41.24 * R_li 35.76 * G_li 18.05 * B_li
Y <- 21.26 * R_li 71.52 * G_li 07.22 * B_li
Z <- 01.93 * R_li 11.92 * G_li 95.05 * B_li
# Convert XYZ to CAM16
## User defined Parameters
X_w <- 96.4212
Y_w <- 100
Z_w <- 82.5188
L_A <- 40
Y_b <- 20
surround <- 2
discounting <- FALSE
## Predefined functions and constants
### M16
M16 <- matrix(c(0.401288,-0.250268,-0.002079,
0.650173, 1.204414, 0.048952,
-0.051461, 0.045854, 0.953127), nrow = 3, ncol=3)
### lerp
lerp <- function(a,b,c){
(1 - c) * a c * b
}
### Crop
crop <- function(a,b,c){
pmin(pmax(c, a), b)
}
### Define adapt
adapt <- function(component){
con <- (F_L * abs(component) * 0.01)^0.42
sign(component) * 400 * con / (con 27.13)
}
### Define unadapt
unadapt <- function(component){
sign(component) * 100 / F_L * ((27.13* abs(component))/(400-abs(component)))^2.38095238095
}
# Calculations
## Calculate "c"
if (surround >=1){
c <- lerp(0.59, 0.69, surround-1)
}else{
c <- lerp(0.525, 0.59, surround)
}
## Calculate "F" and "N_c"
if (c >= 0.59){
N_c <- lerp(0.9, 1.0, (c - 0.59)/.1)
} else {
N_c <- lerp(0.8, 0.9, (c - 0.525)/0.065)
}
## Calculate "k"
k <- 1/(5*L_A 1)
## Calculate F_L
F_L <- k^4 * L_A 0.1 * (1-k^4)^2 * (5 * L_A)^0.33333333333
## Calculate n
n <- Y_b / Y_w
## Calculate z
z <- 1.48 sqrt(n)
## Calculate N_bb
N_bb <- 0.725 * n^-0.2
## Calculate D
if (discounting == FALSE){
D <- crop(0,1,N_c* (1 - 1/3.6 * exp((-L_A - 42)/92)))
}else {
D <- 1
}
## Calculate the "RGB_w"
RGB_w <- matrix(c(M16[1,1] * X_w M16[1,2] * Y_w M16[1,3] * Z_w,
M16[2,1] * X_w M16[2,2] * Y_w M16[2,3] * Z_w,
M16[3,1] * X_w M16[3,2] * Y_w M16[3,3] * Z_w), nrow = 3, ncol=1)
# Calculate the "D_RGB"
D_RGB <- apply(RGB_w, c(1, 2), function(x)((1 - D) * 1 D * Y_w/x))
# Calculate the "D_RGB_inv"
D_RGB_inv <- apply(D_RGB,c(1, 2),function(x)1/x)
# Calculate the "RGB_cw"
RGB_cw <- RGB_w*D_RGB
# Calculate RGB_aw
RGB_aw <- apply(RGB_cw, c(1,2), adapt)
# Calculate A_w
A_w <- N_bb * ( 2 * RGB_aw[1,1] RGB_aw[2,1] 0.05 * RGB_aw[3,1])
# Calculate RGB_a
R_a <- adapt((M16[1,1] * X M16[1,2] * Y M16[1,3] * Z) * D_RGB[1,1])
B_a <- adapt((M16[2,1] * X M16[2,2] * Y M16[2,3] * Z) * D_RGB[2,1])
G_a <- adapt((M16[3,1] * X M16[3,2] * Y M16[3,3] * Z) * D_RGB[3,1])
# Calculate Hue
a <- R_a (-12 * G_a B_a) / 11
b <- (R_a G_a - 2 * B_a) / 9
h_rad <- atan2(b, a)
h_ucs <- h_rad*(180.0/pi)
# Calculate Lightness (J)
e_t <- 0.25 * (cos(h_rad 2) 3.8)
A <- N_bb * (2*R_a G_a 0.05*B_a)
J <- 100 * ((A / A_w)^(c*z))
J_ucs <- 1.7 * J / (1 0.007 * J)
# Calculate brightness (Q)
Q <- 4/c * sqrt(J/100) * (A_w 4) * (F_L^0.25)
# Calculate chroma (C)
t <- (5000 / 13 * N_c * N_bb * e_t * sqrt(a*a b*b)) / (R_a G_a 1.05 * B_a 0.305)
alpha <- t^0.9*(1.64 - 0.29^n)^0.73
C <- alpha * sqrt(J/100)
# Calculate colorfulness (M)
M <- C * F_L^0.25
M_ucs <- log(1 0.0228 * M) / 0.0228
# Calculate redness-greenness(a)
a_ucs <- M * cos(h_rad)
# Calculate yellowness-blueness(b)
b_ucs <- M * sin(h_rad)
# Calculate Saturation (s)
s <- 50 * sqrt(alpha*c /(A_w 4))
return(tibble(R255, G255, B255, h_ucs, J_ucs ,M_ucs ,a_ucs, b_ucs))
}
nc <- 5
rgb_vals <- tibble(r1 = rdunif(nc, b=255, a=0), g1 = rdunif(nc, b=255, a=0), b1 = rdunif(nc, b=255, a=0))
test <- 1
test_df <- tibble(h_ucs = numeric(), J_ucs = numeric(), M_ucs = numeric(), a_ucs = numeric(), b_ucs = numeric())
while(test <= nrow(rgb_vals)){
test_r <- sRGB_to_CAM16UCS(rgb_vals[test, 1],rgb_vals[test, 2],rgb_vals[test, 3])
test_df <- rbind(test_df, test_r)
test <- test 1
}
output <- cbind(rgb_vals, test_df)
print(output)
openxlsx::write.xlsx(output, "rgb2camucs.xlsx")
threw an error as following
Error in atan2(b, a) : non-numeric argument to mathematical function
>
> output <- cbind(rgb_vals, test_df)
Error in data.frame(..., check.names = FALSE) :
arguments imply differing number of rows: 5, 0
> print(output)
Error in print(output) : object 'output' not found
UPDATE: If I wrap the a and b with as.numeric() function, it throws the following error message:
Error:
! Column names `r1`, `r1`, `r1`, `r1`, `r1`, and 1 more must not be duplicated.
Use .name_repair to specify repair.
Caused by error in `repaired_names()`:
! Names must be unique.
x These names are duplicated:
* "r1" at locations 1, 2, 3, 5, 6, etc.
Run `rlang::last_error()` to see where the error occurred.
>
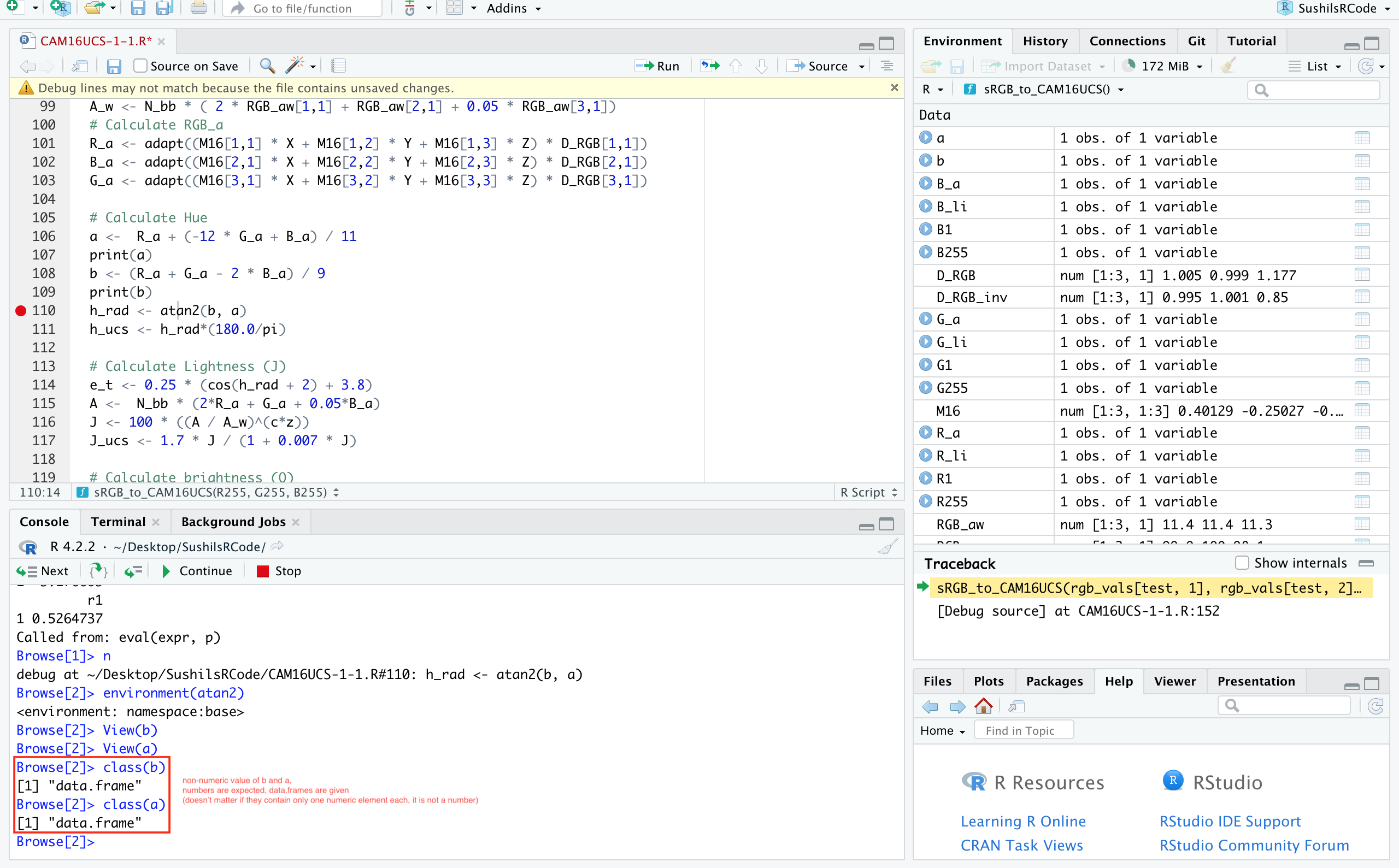
CodePudding user response:
This is because b and a are indeed non-numeric arguments. They are data.frames. Replacing that line with h_rad <- atan2(b$r1, a$r1) makes it work as class(b$r1) results in numeric.
Note that the last line of code where output is exported to an XLSX does not work.
I found this using RStudio's debugger, setting a breakpoint to that line and then entering class(b) into the console on the bottom.