I have 2 data frames like the one below:
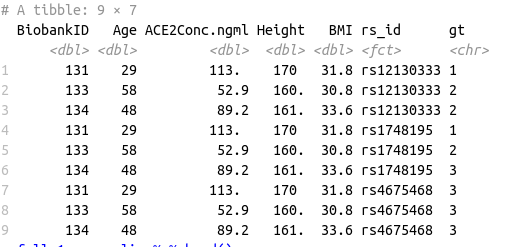
Data Frame 1:
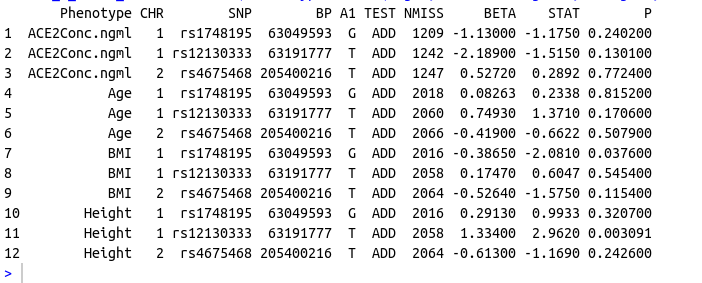
Data Frame 2
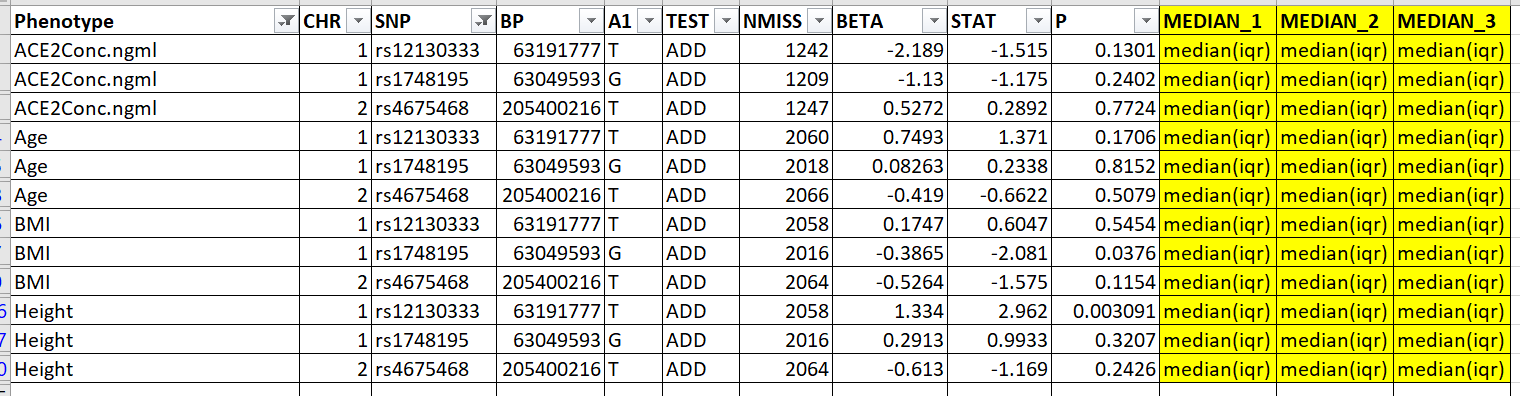
I am trying to get Median(IQR) for the phenotypes (Age, ACE2, Height and BMI) for Data Frame 1 by each rs_id's genotype (gt) and add it to Data Frame 2 by rs_id and Phenotpe. This is the resultant data frame I am expecting:
I am giving the minimal sample dput here.
Data Frame 1:
structure(list(BiobankID = c(131, 133, 134, 136, 140, 141, 143,
144, 145, 131, 133, 134, 136, 140, 141, 143, 144, 145, 131, 133,
134, 136, 140, 141, 143, 144, 145), Age = c(29, 58, 48, 44, 40,
32, 43, 61, 64, 29, 58, 48, 44, 40, 32, 43, 61, 64, 29, 58, 48,
44, 40, 32, 43, 61, 64), ACE2Conc.ngml = c(113.49, 52.91, 89.25,
74.82, 28.5, 97.33, 97.53, 63.72, 56.86, 113.49, 52.91, 89.25,
74.82, 28.5, 97.33, 97.53, 63.72, 56.86, 113.49, 52.91, 89.25,
74.82, 28.5, 97.33, 97.53, 63.72, 56.86), Height = c(170, 160.2,
161.3, 168, 148, 175.2, 167, 174, 163.1, 170, 160.2, 161.3, 168,
148, 175.2, 167, 174, 163.1, 170, 160.2, 161.3, 168, 148, 175.2,
167, 174, 163.1), BMI = c(31.8, 30.8, 33.6, 31.8, 20.1, 26.8,
26, 27.4, 31.3, 31.8, 30.8, 33.6, 31.8, 20.1, 26.8, 26, 27.4,
31.3, 31.8, 30.8, 33.6, 31.8, 20.1, 26.8, 26, 27.4, 31.3), rs_id = structure(c(9L,
9L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 11L, 11L, 11L, 11L, 11L, 11L,
11L, 11L, 11L, 35L, 35L, 35L, 35L, 35L, 35L, 35L, 35L, 35L), .Label = c("rs2278426",
"rs1122326", "rs10033119", "rs4932373", "rs2071410", "rs41303171",
"rs4646116", "rs328", "rs12130333", "rs1748197", "rs1748195",
"rs79566395", "rs55843109", "rs4703642", "rs199347", "rs148110342",
"rs16909225", "rs6814581", "rs3825041", "rs67904839", "rs11508026",
"rs368669", "rs673548", "rs2266788", "rs9457", "rs7770437", "rs1046322",
"rs1997623", "rs964184", "rs6589566", "rs12440118", "rs2160669",
"rs4375003", "rs114060881", "rs4675468", "rs5754701", "rs12642481"
), class = "factor"), gt = c("1", "2", "2", "1", "1", "2", "1",
"1", "1", "1", "2", "3", "1", "2", "2", "1", "2", "1", "3", "3",
"3", "3", "3", "3", "3", "3", "3")), row.names = c(NA, -27L), class = c("tbl_df",
"tbl", "data.frame"))
Data Frame 2
structure(list(Phenotype = c("ACE2Conc.ngml", "ACE2Conc.ngml",
"ACE2Conc.ngml", "Age", "Age", "Age", "BMI", "BMI", "BMI", "Height",
"Height", "Height"), CHR = c(1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L,
2L, 1L, 1L, 2L), SNP = c("rs1748195", "rs12130333", "rs4675468",
"rs1748195", "rs12130333", "rs4675468", "rs1748195", "rs12130333",
"rs4675468", "rs1748195", "rs12130333", "rs4675468"), BP = c(63049593L,
63191777L, 205400216L, 63049593L, 63191777L, 205400216L, 63049593L,
63191777L, 205400216L, 63049593L, 63191777L, 205400216L), A1 = c("G",
"T", "T", "G", "T", "T", "G", "T", "T", "G", "T", "T"), TEST = c("ADD",
"ADD", "ADD", "ADD", "ADD", "ADD", "ADD", "ADD", "ADD", "ADD",
"ADD", "ADD"), NMISS = c(1209L, 1242L, 1247L, 2018L, 2060L, 2066L,
2016L, 2058L, 2064L, 2016L, 2058L, 2064L), BETA = c(-1.13, -2.189,
0.5272, 0.08263, 0.7493, -0.419, -0.3865, 0.1747, -0.5264, 0.2913,
1.334, -0.613), STAT = c(-1.175, -1.515, 0.2892, 0.2338, 1.371,
-0.6622, -2.081, 0.6047, -1.575, 0.9933, 2.962, -1.169), P = c(0.2402,
0.1301, 0.7724, 0.8152, 0.1706, 0.5079, 0.0376, 0.5454, 0.1154,
0.3207, 0.003091, 0.2426)), class = "data.frame", row.names = c(NA,
-12L))
Can someone please help?
CodePudding user response:
A tidyverse approach:
library(tidyverse)
df1 %>%
#for each id and gt, take the median of each variable:
group_by(rs_id, gt) %>%
summarise(across(c(Age, ACE2Conc.ngml, Height, BMI), median)) %>%
ungroup() %>% #stop doing operations for each id and gt
#change to a Phenotype | value format, instead of one column for each variable
pivot_longer(c(Age, ACE2Conc.ngml, Height, BMI), names_to = "Phenotype") %>%
#change to a Phenotype | median_1 | median_2 | median_3
pivot_wider(values_from = value, names_from = gt, names_prefix = "MEDIAN_") %>%
#merge and arrange the rows orders
rename(SNP = rs_id) %>%
merge(df2) %>%
arrange(Phenotype, SNP)
Result:
SNP Phenotype MEDIAN_1 MEDIAN_2 MEDIAN_3 CHR BP A1 TEST NMISS BETA STAT P
1 rs12130333 ACE2Conc.ngml 69.270 89.250 NA 1 63191777 T ADD 1242 -2.18900 -1.5150 0.130100
2 rs1748195 ACE2Conc.ngml 86.175 58.315 89.25 1 63049593 G ADD 1209 -1.13000 -1.1750 0.240200
3 rs4675468 ACE2Conc.ngml NA NA 74.82 2 205400216 T ADD 1247 0.52720 0.2892 0.772400
4 rs12130333 Age 43.500 48.000 NA 1 63191777 T ADD 2060 0.74930 1.3710 0.170600
5 rs1748195 Age 43.500 49.000 48.00 1 63049593 G ADD 2018 0.08263 0.2338 0.815200
6 rs4675468 Age NA NA 44.00 2 205400216 T ADD 2066 -0.41900 -0.6622 0.507900
7 rs12130333 BMI 29.350 30.800 NA 1 63191777 T ADD 2058 0.17470 0.6047 0.545400
8 rs1748195 BMI 31.550 27.100 33.60 1 63049593 G ADD 2016 -0.38650 -2.0810 0.037600
9 rs4675468 BMI NA NA 30.80 2 205400216 T ADD 2064 -0.52640 -1.5750 0.115400
10 rs12130333 Height 167.500 161.300 NA 1 63191777 T ADD 2058 1.33400 2.9620 0.003091
11 rs1748195 Height 167.500 167.100 161.30 1 63049593 G ADD 2016 0.29130 0.9933 0.320700
12 rs4675468 Height NA NA 167.00 2 205400216 T ADD 2064 -0.61300 -1.1690 0.242600
CodePudding user response:
Not a real answwer, but may me a helpful hint: I've noticed that you work with data.frames which are also tibble so you might be a tidyverse-inhabitant. Did you consider using tidyr::gather resp. tidy::spread? and dplyr::summarize https://dplyr.tidyverse.org/reference/summarise.html e.g. useful functions ... IQR() ...