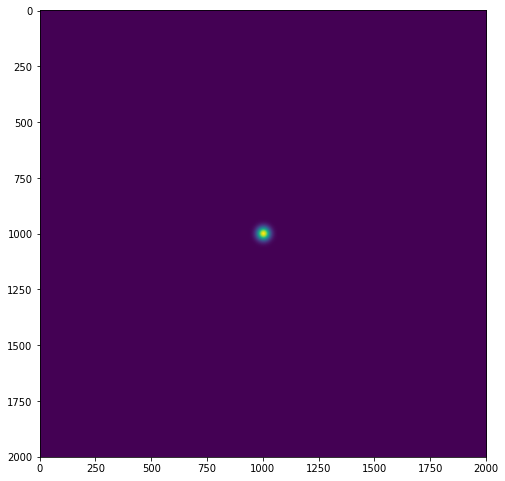
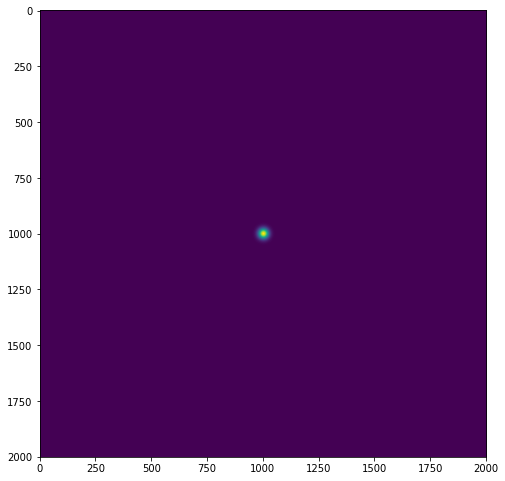
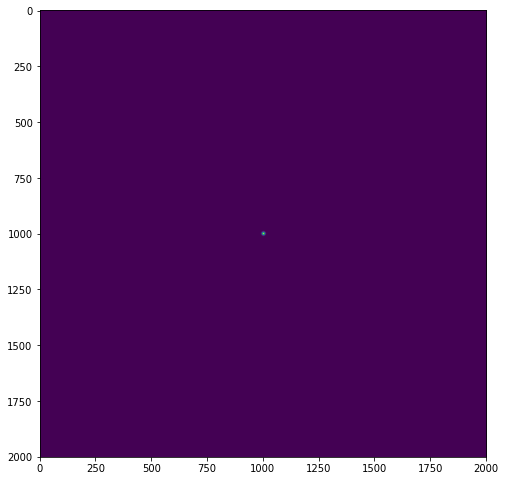
We know the Fourier transform of a Gaussian filter is again Gaussian in the frequency domain, I have written the following method to build a Gaussian kernel:
def get_gaussian(size, sigma):
g_kernel = np.zeros((size,size))
x_center = size // 2
y_center = size // 2
for i in range(size):
for j in range(size):
g_kernel[i,j] = np.exp(-((i - x_center)**2 (j - y_center)**2) / (2*sigma**2))
return 1/(2*np.pi * sigma**2)* g_kernel
plt.figure(figsize=(8, 10))
sigma = 20
g_spatial = get_gaussian(img2.shape[0],sigma)
plt.imshow(g_spatial)
plt.figure(figsize=(8, 10))
g_frequnecy = fft.fftshift(fft.fft2(g_spatial))
plt.imshow(np.abs(g_frequnecy))
plt.figure(figsize=(8, 10))
g_spatial = get_gaussian(img2.shape[0],sigma / (np.sqrt(2) * np.pi))
plt.imshow(g_spatial)
The second and third Gaussians don't seem to match up at all. I will attach the pictures of all three Gaussian kernels below. I'm pretty sure I have calculated the variance of each Gaussian kernel such that they would match based on the 2D Fourier transform of a Gaussian kernel. Where have I made a mistake?
CodePudding user response:
In the frequency domain, the Gaussian has a sigma of size / (2 * pi * sigma), with size the size of the image, and sigma the spatial-domain sigma. Yes, for a non-square image, an isotropic Gaussian in the spatial domain is not isotropic in the frequency domain.
Your computation sigma / (np.sqrt(2) * np.pi) is wrong. One quick way to tell it's not correct is that a larger sigma in the spatial domain also produces a larger sigma in the frequency domain using your computation. But we know that scaling up (==making larger) in the spatial domain scales the frequency domain down, as larger wavelengths have smaller frequencies.