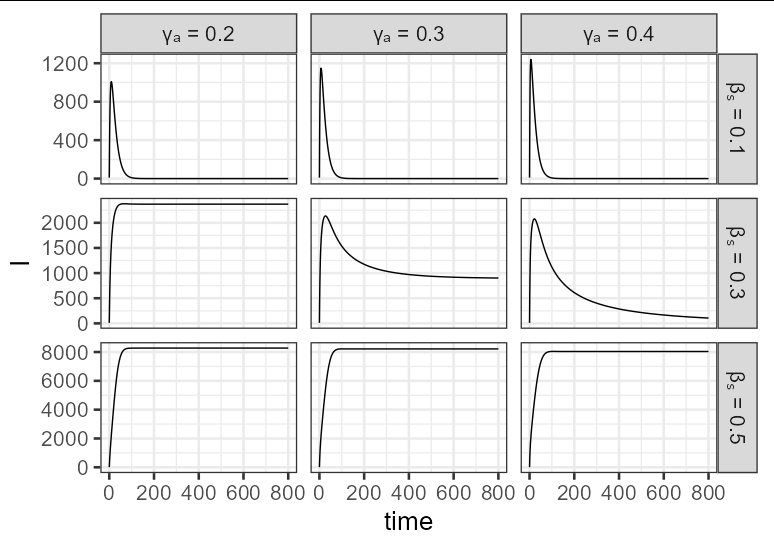
I have been trying to plot a graph that looks like the one given below for a model that I saw on a paper.
The R codes for the model are as follows. I want to draw a plot that looks like the above for beta_s = c(0.1, 0.3, 0.5) and gamma_a = c(0.2, 0.3, 0.4) over time = 600 days.
library(deSolve)
initial_values <- c(S = 4000, A = 3000, I = 10, U = 1000)
parameters <- c(theta = 2500, beta_s = 0.274, gamma_a = 0.29, alpha_a = 0.4775, alpha_u
= 0.695, mu = 0.062, q_i = 0.58, eta_i = 0.009, eta_u = 0.05)
time_series <- 0:800
disease_model <- function(time, state, parameters) {
with(as.list(c(state, parameters)), {
N = S A I U
dS <- theta - ((beta_s*S)/N)*(alpha_a*A alpha_u*U) - mu*S
dA <- ((beta_s*S)/N)*(alpha_a*A alpha_u*U) - gamma_a*A - mu*A
dI <- q_i*gamma_a*A - eta_i*I - mu*I
dU <- gamma_a*(1 - q_i)*A - eta_u*U - mu*U
return(list(c(dS, dA, dI, dU)))
})
}
model <- ode(initial_values, time_series, disease_model, parameters)
summary(model)
matplot(model, type="l", lty=1, main="Disease model", xlab="Time")
legend <- colnames(model)[2:5]
legend("right", legend=legend, col=2:5, lty = 1)
Any support would be greatly appreciated. Thank you!
CodePudding user response:
It looks like this was made in ggplot. You would have to create one data frame for each combination of the beta and gamma values you wanted, then bind them together. You can then use facting to achieve the end result. I presume it is column I you want to plot:
df <- do.call(rbind, Map(function(beta, gamma) {
parameters <- c(theta = 2500, beta_s = beta, gamma_a = gamma,
alpha_a = 0.4775, alpha_u
= 0.695, mu = 0.062, q_i = 0.58,
eta_i = 0.009, eta_u = 0.05)
res <- ode(initial_values, time_series, disease_model, parameters)
res <- as.data.frame(res)
res$beta <- paste('\u03b2\u209b =', beta)
res$gamma <- paste('\u03b3\u2090 =', gamma)
res},
beta = rep(c(0.1, 0.3, 0.5), 3),
gamma = rep(c(0.2, 0.3, 0.4), each = 3)))
library(ggplot2)
ggplot(df, aes(time, I))
geom_line()
facet_grid(beta~gamma, scales = 'free_y')
theme_bw(base_size = 20)