I have the following data frame:
table <- data.frame(pop_1 = c("AL","AL","AL","AL","AL","AL","AL","ALT","ALT","ALT","ALT","ALT","ALT","BU","BU","BU","BU","BU","IRK","IRK","IRK","IRK","KK","KK","KK","KYA","KYA","TU"),
pop_2 = c("ALT","BU","IRK","KK","KYA","TU","ZAB","BU","IRK","KK","KYA","TU","ZAB","IRK","KK","KYA","TU","ZAB","KK","KYA","TU","ZAB","KYA","TU","ZAB","TU","ZAB","ZAB"),
value = c(0.43447,0.15267,0.25912,0.10435,0.19238,0.19186,0.18155,0.34969,0.07506,0.29206,0.13597,0.46354,0.17870,0.18658,0.02297,0.08851,0.18950,0.05176,0.12086,0.02690,0.29669,0.05551,0.04910,0.15779,0.03276,0.23422,0.00568,0.22181))
How to convert it to an asymmetric matrix with empty (or NA, etc.) cells like this:
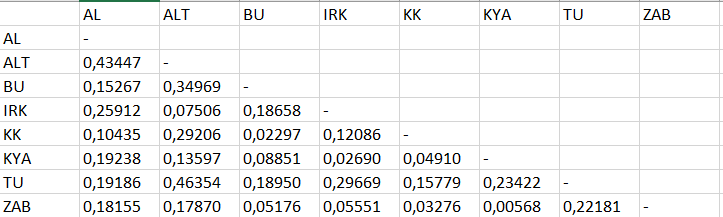
CodePudding user response:
Minor change to your data frame, extra "AL", "AL", "NA" combination at the start. You'll want to do the same for an extra "ZAB" at the end:
df<- data.frame(pop_1 = c("AL","AL","AL","AL","AL","AL","AL","AL","ALT","ALT","ALT","ALT","ALT","ALT","BU","BU","BU","BU","BU","IRK","IRK","IRK","IRK","KK","KK","KK","KYA","KYA","TU"),
pop_2 = c("AL","ALT","BU","IRK","KK","KYA","TU","ZAB","BU","IRK","KK","KYA","TU","ZAB","IRK","KK","KYA","TU","ZAB","KK","KYA","TU","ZAB","KYA","TU","ZAB","TU","ZAB","ZAB"),
value = c(NA,0.43447,0.15267,0.25912,0.10435,0.19238,0.19186,0.18155,0.34969,0.07506,0.29206,0.13597,0.46354,0.17870,0.18658,0.02297,0.08851,0.18950,0.05176,0.12086,0.02690,0.29669,0.05551,0.04910,0.15779,0.03276,0.23422,0.00568,0.22181))
library(tidyverse)
pivot_wider(df, names_from=pop_1, values_from=value)
pop_2 AL ALT BU IRK KK KYA TU
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 AL NA NA NA NA NA NA NA
2 ALT 0.434 NA NA NA NA NA NA
3 BU 0.153 0.350 NA NA NA NA NA
4 IRK 0.259 0.0751 0.187 NA NA NA NA
5 KK 0.104 0.292 0.0230 0.121 NA NA NA
6 KYA 0.192 0.136 0.0885 0.0269 0.0491 NA NA
7 TU 0.192 0.464 0.190 0.297 0.158 0.234 NA
8 ZAB 0.182 0.179 0.0518 0.0555 0.0328 0.00568 0.222
CodePudding user response:
Create a vector of all the unique values in the pop_1 and pop_2 columns of the data frame. This will be the names of the rows and columns of the matrix.
populations <- unique(c(table$pop_1, table$pop_2))
Create an empty matrix with the same number of rows and columns as the vector from step 1, using the matrix function. Set the default value of the matrix to NA using the value argument.
matrix <- matrix(NA, nrow = length(populations), ncol = length(populations))
Use the rownames and colnames functions to set the names of the rows and columns of the matrix to the values in the populations vector.
rownames(matrix) <- populations
colnames(matrix) <- populations
Use a for loop to iterate over the rows of the data frame. For each row, use the pop_1 and pop_2 columns to find the corresponding cells in the matrix, and use the value column to set the value of those cells.
for (i in 1:nrow(table)) {
row_name <- table[i, "pop_1"]
col_name <- table[i, "pop_2"]
value <- table[i, "value"]
matrix[row_name, col_name] <- value
}
After these steps, the matrix should be an asymmetric matrix with the values from the data frame in the appropriate cells, and NA in all the other cells.
When you look at the results of matrix:
AL ALT BU IRK KK KYA TU ZAB
AL NA 0.43447 0.15267 0.25912 0.10435 0.19238 0.19186 0.18155
ALT NA NA 0.34969 0.07506 0.29206 0.13597 0.46354 0.17870
BU NA NA NA 0.18658 0.02297 0.08851 0.18950 0.05176
IRK NA NA NA NA 0.12086 0.02690 0.29669 0.05551
KK NA NA NA NA NA 0.04910 0.15779 0.03276
KYA NA NA NA NA NA NA 0.23422 0.00568
TU NA NA NA NA NA NA NA 0.22181
ZAB NA NA NA NA NA NA NA NA
