I am trying to plot a dose-response curve (for degradation) using R. I would like to get the interpolated X-axis value (dose concentration value) at the selected percent substrate remaining value (i.e., Y value, for example, Y = 50) as could be interpolated in GraphPad PRISM.
I used the code given below to predict the EC50 at the inflection point. The EC50 value obtained using R and GraphPad PRISM is the same for EC50 at the inflection point. Using GraphPad PRISM, I am able to get the interpolated X-axis (dose concentration) value at Y = 50 as 195.391 nM. However, using R I am not able to get the X-axis value at the selected Y-axis value (Y = 50).
However, I could not do so using R. The value I get with ED() function in drc package is the same as the EC50 value at the inflection point as shown in the code below. The response values for the different doses of degrader are normalized to 100% based on DMSO values. Can someone let me know what am I missing?
EC50 at inflection point and EC50 at 50% (Y = 50) using R
library(drc)
df <- data.frame(
x = c(1,3.2,10,32,100,320,1000,3200,10000113.891531792259),
y = c(108.505,119.305,98.01,89.275,65.305,39.87,21.95,7.32,8.695))
model <- drm(y~x, data=df, fct=LL.4(names = c("Slope", "Lower Limit", "Upper Limit", "EC50 at IP")))
plot(model, type="all", col = "blue", ylab = "% Remaining", xlab = "Concentration (nM)")
axis(2, at = seq(0, 120, 5), labels = F)
axis(1, at = seq(100, 1000, 100), labels = F)
summary(model)
Model fitted: Log-logistic (ED50 as parameter) (4 parms)
Parameter estimates:
Estimate Std. Error t-value p-value
Slope:(Intercept) 0.82802 0.16507 5.0160 0.004049 **
Lower Limit:(Intercept) 3.47712 6.15672 0.5648 0.596627
Upper Limit:(Intercept) 115.18939 5.38586 21.3874 0.000004144 ***
EC50 at IP:(Intercept) 129.99815 32.25388 4.0305 0.010017 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error:
5.507845 (5 degrees of freedom)
ED(model, 50)
Estimated effective doses
Estimate Std. Error
e:1:50 129.998 32.254
CodePudding user response:
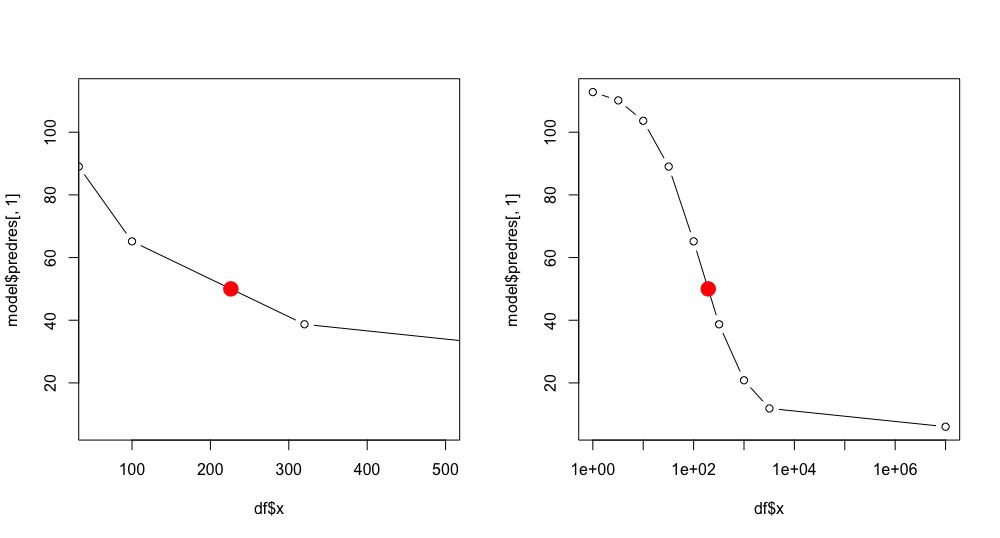
You can use the approx function to do linear interpolation. I used this function to do linear interpolation using your x variable as well as the log of your x variable. Interpolating with the log works visually when plotting on a log scale (as you do) and the interpolating with the level works when plotting in levels. I get somewhat different values: 226.1137 in levels and 194.7934 in logs. Here is my code:
summary(model)
par(mfrow = c(1, 2))
answer <- approx(model$predres[, 1], df$x, 50)
answer
plot(df$x, model$predres[, 1], typ = "b", xlim = c(50, 500))
points(answer[[2]], answer[[1]], col = "red", pch = 19, cex = 2)
answer2 <- approx(model$predres[, 1], log(df$x), 50)
exp(answer2$y)
plot(df$x, model$predres[, 1], typ = "b", log = "x")
points(exp(answer2[[2]]), answer[[1]], col = "red", pch = 19, cex = 2)
hope that is helpful.
CodePudding user response:
My bad! I overlooked the ED function. I should have simply used ED() function with type = 'absolute'. Thanks to this post: Inverse prediction using drm package in R for the vital clue.
ED(object = model, respLev = 50, type = c("absolute"))
Estimate Std. Error
e:1:50 195.379 52.166
ED(object = model, respLev = 50, type = c("relative"))
Estimate Std. Error
e:1:50 129.998 32.254