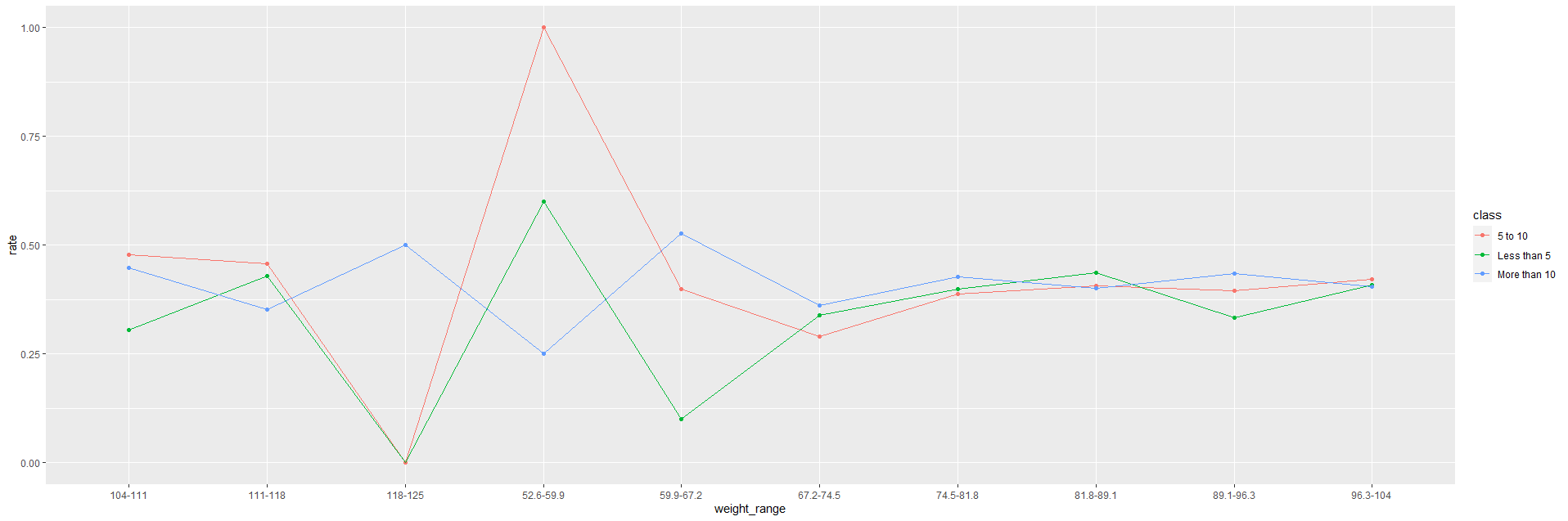
I am working with the R programming language.
I simulated a dataset, did some data manipulation (in a very clumsy way) and made the following plot.
Simulate Data:
# Step 1 : Simulate Data
set.seed(123)
Hospital_Visits = sample.int(20, 5000, replace = TRUE)
Weight = rnorm(5000, 90, 10)
disease <- c("Yes","No")
disease <- sample(disease, 5000, replace=TRUE, prob=c(0.4, 0.6))
Disease <- as.factor(disease)
my_data = data.frame(Weight, Hospital_Visits, Disease)
my_data$hospital_ntile <- cut(my_data$Hospital_Visits, breaks = c(0, 5, 10, Inf), labels = c("Less than 5", "5 to 10", "More than 10"), include.lowest = TRUE)
Data Manipulation:
# Step 2: Data Manipulation:
my_data$weight_ntile <- cut(my_data$Weight, breaks = seq(min(my_data$Weight), max(my_data$Weight), by = (max(my_data$Weight) - min(my_data$Weight)) / 10), include.lowest = TRUE)
# Create a dataset for rows where hospital_ntile = 'Less than 5'
df1 <- subset(my_data, hospital_ntile == "Less than 5")
# Create a dataset for rows where hospital_ntile = '5 to 10'
df2 <- subset(my_data, hospital_ntile == "5 to 10")
# Create a dataset for rows where hospital_ntile = 'More than 10'
df3 <- subset(my_data, hospital_ntile == "More than 10")
avg_disease_rate_df1 <- tapply(df1$Disease == "Yes", df1$weight_ntile, mean)
avg_disease_rate_df2 <- tapply(df2$Disease == "Yes", df2$weight_ntile, mean)
avg_disease_rate_df3 <- tapply(df3$Disease == "Yes", df3$weight_ntile, mean)
avg_disease_rate_df1[is.na(avg_disease_rate_df1)] <- 0
avg_disease_rate_df2[is.na(avg_disease_rate_df2)] <- 0
avg_disease_rate_df3[is.na(avg_disease_rate_df3)] <- 0
#transform into dataset
names = names(avg_disease_rate_df1)
rate_1 = as.numeric(avg_disease_rate_df1)
rate_2 = as.numeric(avg_disease_rate_df2)
rate_3 = as.numeric(avg_disease_rate_df3)
# stack data
d1 = data.frame(class = "Less than 5", names = names, rate = rate_1)
d2 = data.frame(class = "5 to 10", names = names, rate = rate_2)
d3 = data.frame(class = "More than 10", names = names, rate = rate_3)
plot_data = rbind(d1, d2, d3)
Make Plot:
library(ggplot2)
ggplot(plot_data, aes(x=names, y=rate, group = class, color=class)) geom_point() geom_line() theme_bw()
For some reason, the ordering on the x-axis is not in order - right now it appears randomly, I would like to make it from smallest to largest.
I consulted some references which showed how to change this manually - but is there some option within ggplot2 that allows for this ordering to be automatically corrected?
Thanks!
CodePudding user response:
EDIT - you can either do this at the 1) plotting step or at the 2) data manipulation step.
1) - Option for Doing it at the Plotting Step
I think the easiest way to do this is to turn your x-axis variable into a factor and order it.
Right now it's a character
str(plot_data)
'data.frame': 30 obs. of 3 variables:
$ class: chr "Less than 5" "Less than 5" "Less than 5" "Less than 5" ...
$ names: chr "[52.6,59.9]" "(59.9,67.2]" "(67.2,74.5]" "(74.5,81.8]" ...
$ rate : num 0.6 0.1 0.339 0.399 0.438 ...
So you can make it into a factor and then check the levels:
plot_data$names <- as.factor(plot_data$names)
levels(plot_data$names)
which will show them in a somewhat random order:
[1] "(104,111]" "(111,118]" "(118,125]" "(59.9,67.2]" "(67.2,74.5]" "(74.5,81.8]" "(81.8,89.1]" "(89.1,96.3]" "(96.3,104]"
[10] "[52.6,59.9]"
You can re-level them then with the forcats library (there are other options but I like this one):
plot_data$names <- fct_relevel(plot_data$names,
c("[52.6,59.9]", "(59.9,67.2]", "(67.2,74.5]",
"(74.5,81.8]", "(81.8,89.1]", "(89.1,96.3]",
"(96.3,104]", "(104,111]", "(111,118]" ))
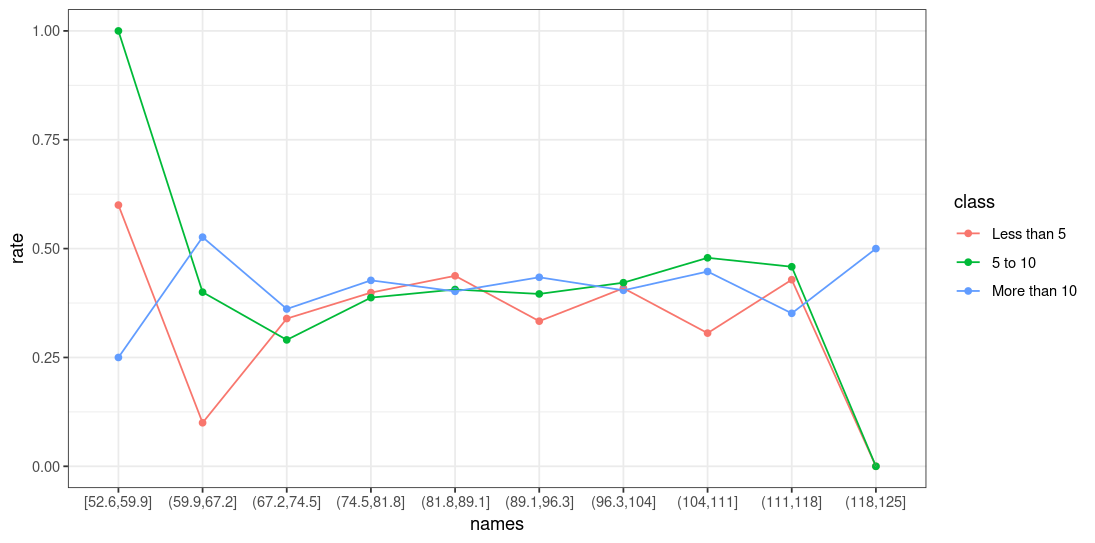
And then your plot will look like this:
ggplot(plot_data, aes(x=names, y=rate, group = class, color=class))
geom_point()
geom_line()
theme_bw()
2) - Option for Doing it at the Data Manipulation Step
When you make weight_ntile you use cut() to which you can ask for an ordered result (ordered_result = TRUE) which will get you your ordered factor. But then the rest of the data manipulation will get rid of that ordered factor if left as is. Instead, you could do it all in a pipe series with dplyr and tidyr. Here's a way to do that:
my_data$weight_ntile <- cut(my_data$Weight,
breaks = seq(min(my_data$Weight),
max(my_data$Weight),
by = (max(my_data$Weight) - min(my_data$Weight)) / 10),
include.lowest = TRUE,
ordered_result = TRUE)
plot_data <- my_data %>%
# group by your variables of interest
dplyr::group_by(hospital_ntile, weight_ntile, Disease) %>%
# find number of obs for eacvh group
dplyr::summarize(
count = n()
) %>%
# move these from long to wide for easier calculation
tidyr::pivot_wider(
names_from = "Disease", values_from = "count"
) %>%
dplyr::rowwise() %>%
# make the rate calculation, na.rm = TRUE is important for the edge case of
# only one observation (gives the 1.00 rate for 52.6 and 5 to 10)
dplyr::summarize(
rate = Yes / sum(Yes, No, na.rm = TRUE)
) %>%
# need to ungrouop to use complete()
ungroup() %>%
# use complete to fill in what would be NAs, then replace with zeros
# instead of NA's (that's the fill argument), this replaces your
# `avg_disease_rate_df1[is.na(avg_disease_rate_df1)] <- 0` etc step
tidyr::complete(., hospital_ntile, weight_ntile, fill = list(rate = 0)) %>%
# rename to what you use for the plotting step
dplyr::rename(
class = hospital_ntile, names = weight_ntile
)
And then the plot can be made the same way:
ggplot(plot_data, aes(x=names, y=rate, group = class, color=class))
geom_point()
geom_line()
theme_bw()