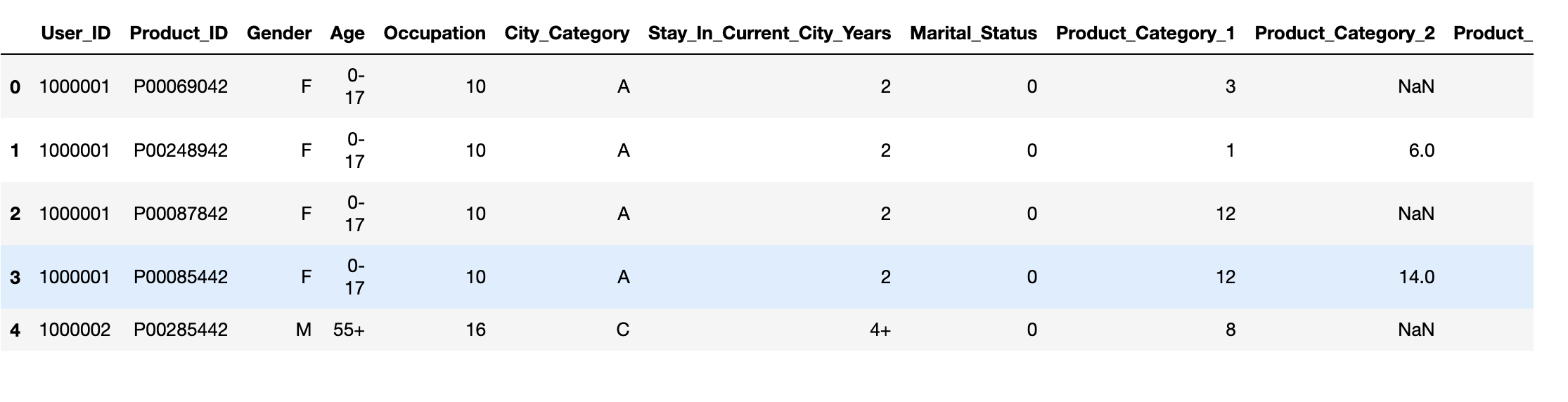
I'm trying to replace the categorical variable in the Gender column - M, F with 0, 1. However, after running my code I'm getting NaN in place of 0 & 1.
Code-
df['Gender'] = df['Gender'].map({'F':1, 'M':0})
Dataframe after running the code-
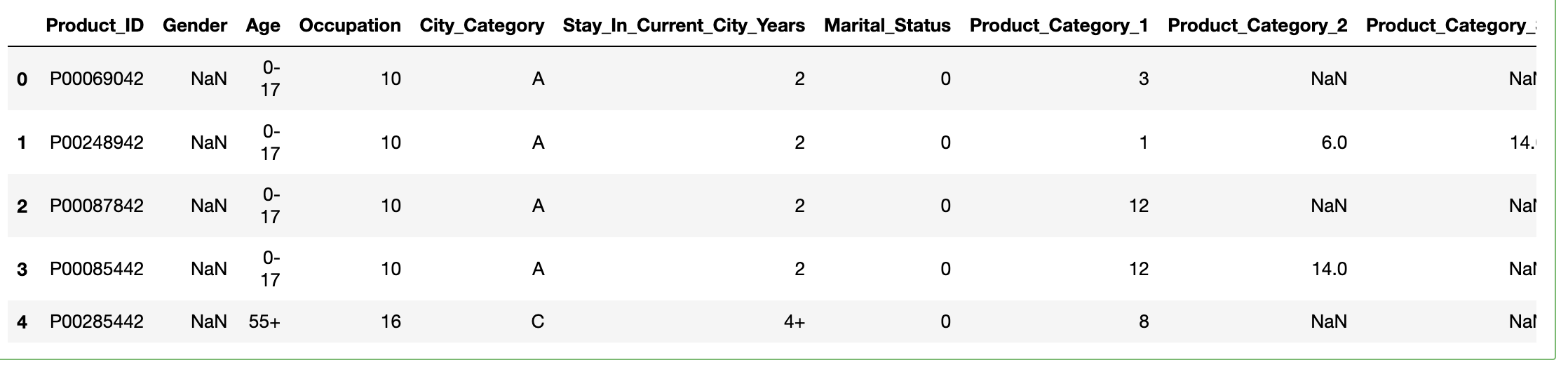
Kindly, suggest a way out!
CodePudding user response:
If you have whitespace in your strings, then you'll need to strip it out before mapping:
df['Gender'] = df['Gender'].str.strip().map({'F':1, 'M':0})
Instead of using map, you can also check if the string is equal to 'F' and convert that to an integer:
df['Gender'] = (df['Gender'].str.strip() == 'F').astype(int)
For example, with the following df:
Gender Name
0 M Person 0
1 F Person 1
2 F Person 2
3 M Person 3
4 F Person 4
5 M Person 5
6 F Person 6
7 M Person 7
8 F Person 8
9 F Person 9
10 M Person 10
Both above statements give the following result:
0 0
1 1
2 1
3 0
4 1
5 0
6 1
7 0
8 1
9 1
10 0
Name: Gender, dtype: int64
But on my computer the map statement takes ~1.7x the time that the == 'F' statement does:
%timeit x = (df['Gender'].str.strip() == 'F').astype(int)
494 µs ± 84.9 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
%timeit y = df['Gender'].str.strip().map({'F':1, 'M':0})
840 µs ± 2.61 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
CodePudding user response:
Maybe values in your dataframe are different from the expected strings 'F' and 'M'. Try to use LabelEncoder from SkLearn.
from sklearn.preprocessing import LabelEncoder
df['Gender'] = LabelEncoder().fit_transform(df['Gender'])