All,
I've been struggling to change the legend text of my plots to have the varietal epithet ("paludicola" or "lanceolate") italicized while leaving the abbreviation for variety ("var.") unchanged. I've tried using expression and paste, but have never been successful. Sometimes I get an error when I try solutions I've found online, but usually the code runs and nothing changes. Any help with this would be greatly appreciated.
Please don't hesitate to ask clarifying questions. Thanks!
Note that I changed the species names in "species" to varieties, but did not adjust the name of that categorical variable accordingly. Sorry for any confusion that may cause. Specific locations were removed from this version of the data before posting; the original data can't be shared freely online.
The dput() data...
structure(list(sample = c("SW001", "SW002", "SW003", "SW004",
"SW005", "SW006"), species = c("var. lanceolata", "var. lanceolata",
"var. lanceolata", "var. lanceolata", "var. paludicola", "var. paludicola"
), sample_type = c("Voucher, only inflorscence", "Voucher, only inflorscence",
"Voucher, only inflorscence", "Voucher, only inflorscence", "Voucher, entire plant, only leaves and roots",
"Voucher, entire plant, only leaves and roots"), date = c("6/11/22",
"6/11/22", "6/11/22", "6/12/22", "6/2/22", "6/2/22"), GPS = c(NA,
NA, "SALABRD001", NA, "SALAP001", "SALAP001"), latitude = c(NA,
NA, "xx° xx.xxx'", NA, "xx° xx.xxx'", "xx° xx.xxx'"), longitude = c(NA,
NA, "xxx° xx.xxx'", NA, "xxx° xx.xxx'", "xxx° xx.xxx'"), flowering = c("Y",
"Y", "Y", "Y", "N", "N"), notes = c("Recently mowed site", "Recently mowed site",
"Blooming plants around, not as many as xxxxxx. Taller grass, wetter habitat. About 50 plants",
"Patch of 200 plants", "Under Acer rubrum that had died and resprouted. The biggest clump. We've collected seeds from this clump in the past. About 50 plants",
"Under Acer rubrum that had died and resprouted. The biggest clump. We've collected seeds from this clump in the past. About 50 plants"
), tissue = c("Y", "Y", "Y", "Y", "Y", "Y"), habitat = c("Roadside",
"Roadside", "Roadside", "Roadside", "Swamp Strand/Private Property",
"Swamp Strand/Private Property"), location = c("xxxxxx, xxxx",
"xxxxxx, xxxx", "xxxxxx, xxxx", "xxxxxx, xxxx",
"xxxxxx, xxxx",
"xxxxxx, xxxx"
), county = c("Martin", "Martin", "Martin", "Highland", "Palm Beach",
"Palm Beach"), new_lat = c(x, x, xx.xxxxxxxxxxxxx, x, xx.xxxxxxxxxxxxx,
xx.xxxxxxxxxxxxx), new_lon = c(x, x, -xx.xxxxxxxxxxxxx, x, -xx.xxxxxxxxxxxxx,
-xx.xxxxxxxxxxxxx)), row.names = c(NA, -6L), class = c("tbl_df",
"tbl", "data.frame"))
One of my plots...
sacoila_bar<-ggplot(data=sacoila_data, aes(x=location, fill = species))
geom_bar() coord_flip() scale_y_continuous(name="Samples")
scale_x_discrete(name="")
scale_fill_manual(values=c("#A9C0AA", "#BF90AC"))
theme(plot.background = element_rect(fill = "#FFFFFF"), panel.background = element_rect(fill = "FFFFFF"))
labs(fill='Taxon')
ggplotly(sacoila_bar)
Among other things, I've tried
scale_color_manual(labels = c(expression(atop("var", italic("paludicola")))),
(expression(atop("var", italic("lanceolata")))))
and
scale_color_manual(labels = c(expression(paste("var", italic("paludicola")))),
(expression(atop("var", italic("lanceolata"))))).
Both of these methods give me (var. lanceolata,1) and (var. paludicola,1) as my legend text. I know I've tried other code, but I think I must've deleted it and can't remember exactly what it was.
CodePudding user response:
Depending on whether you want the right output in the ggplot or the plotly object, there are two different ways to do it. First, if you just want the right output in the ggplot, you can use:
s <- structure(list(species = c("var. lanceolata", "var. lanceolata",
"var. lanceolata", "var. lanceolata", "var. paludicola", "var. paludicola"
), location = c("xxxxxx, xxxx", "xxxxxx, xxxx", "xxxxxx, xxxx",
"xxxxxx, xxxx", "xxxxxx, xxxx", "xxxxxx, xxxx")), row.names = c(NA,
-6L), class = c("tbl_df", "tbl", "data.frame"))
library(ggplot2)
library(plotly)
g1 <- ggplot(data=s, aes(x=location, fill = species))
geom_bar()
coord_flip()
scale_y_continuous(name="Samples")
scale_x_discrete(name="")
scale_fill_manual(values=c("#A9C0AA", "#BF90AC"), labels = c(expression(var.~italic(paludicola)),
expression(var.~italic(lanceolata))))
theme(plot.background = element_rect(fill = "#FFFFFF"), panel.background = element_rect(fill = "#FFFFFF"))
labs(fill='Taxon')
g1
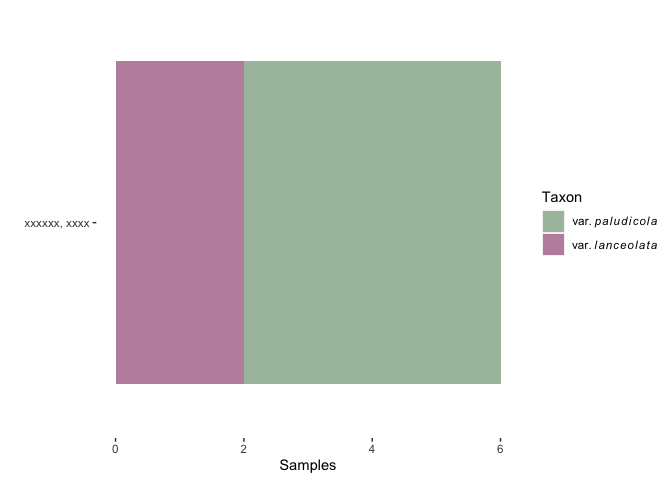
Unfortunately, this doesn't persist when using ggplotly() because that function uses the levels of the fill variable regardless of changes via the scale_XX_manual() functions. This post reminded me that plotly will render HTML, so you can change the levels of your factor variable to have the appropriate HTML tags for italics and while that will look bad in the ggplot, they render correctly via ggplotly().
s$species <- factor(s$species)
levels(s$species) <- c("var. <i>lanceolata</i>", "var. <i>paludicola</i>")
g2 <- ggplot(data=s, aes(x=location, fill = species))
geom_bar()
coord_flip()
scale_y_continuous(name="Samples")
scale_x_discrete(name="")
scale_fill_manual(values=c("#A9C0AA", "#BF90AC"))
theme(plot.background = element_rect(fill = "#FFFFFF"), panel.background = element_rect(fill = "#FFFFFF"))
labs(fill='Taxon')
ggplotly(g2)
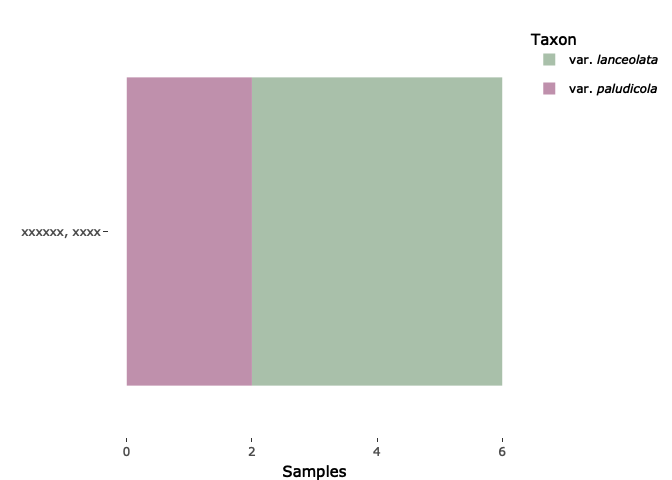
Created on 2023-01-13 by the reprex package (v2.0.1)
