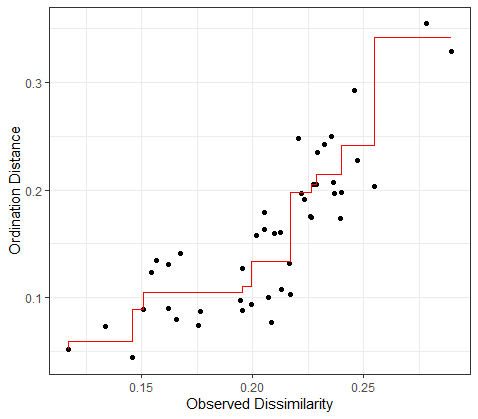
There seems to be quite a bit of information for plotting NMDS outputs (i.e. NMDS1 vs NMDS1) using ggplot2 however I cannot find a way to plot the vegan::stressplot() (shepard's plot) using ggplot2.
Is there a way to produce a ggplot2 version of a metaMDS output?
Reproducible code
library(vegan)
set.seed(2)
community_matrix = matrix(
sample(1:100,300,replace=T),nrow=10,
dimnames=list(paste("community",1:10,sep=""),paste("sp",1:30,sep="")))
example_NMDS=metaMDS(community_matrix, k=2)
stressplot(example_NMDS)