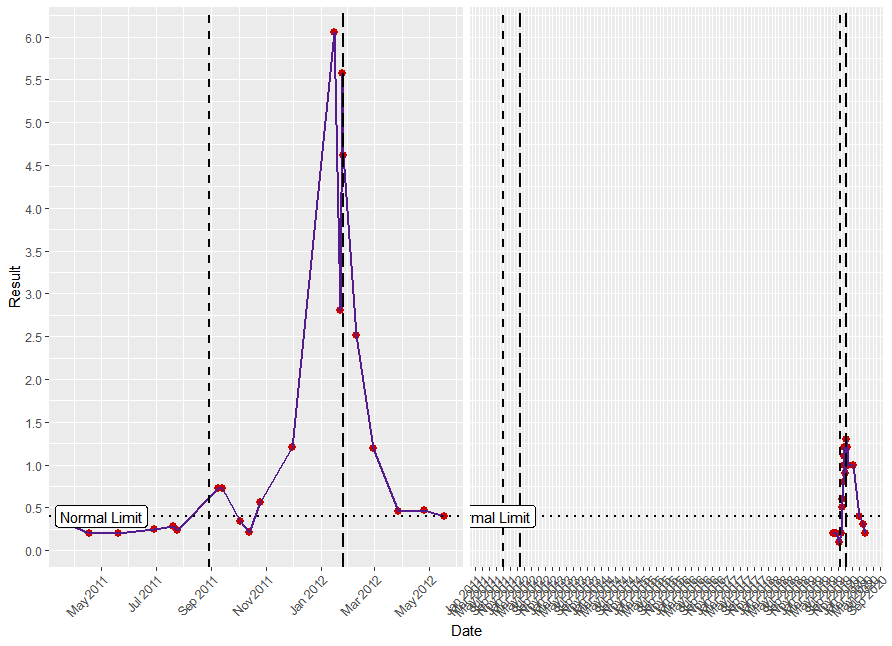
I am creating a plot of y=results x=Date. There is a large period of time that I do not want to plot so have split the plot into 2 facets, excluding the dates not needed.
I want a horizontal line over both facets, but the label "Normal limit" only on the first.
Is this possible?
df = data%>%
filter((Date >= dmy("25/03/2011") & Date <= dmy("18/05/2012")) | #filter unwanted data
(Date >= dmy("18/07/2019") & Date <= dmy("28/04/2020") ))%>%
mutate(bin = Date >= dmy("18/07/2019")) # define the cut point where I want data to be shown
plot = ggplot(df, aes(Date, Result))
geom_point(colour = "red3", size = 2.5)
geom_line( colour= "purple4", size = 1)
facet_wrap(bin ~ ., scale = 'free_x')
scale_x_date(date_labels = "%b %Y",guide = guide_axis(angle = 45), date_breaks = "2 months")
scale_y_continuous(breaks = seq(0,6.2, by=0.5))
theme(strip.text.x = element_blank()) # this removes the true/false bin identification
geom_vline(xintercept = dmy("29/8/2011"), linetype="dashed",
color = "black", size=1, )
geom_vline(xintercept = dmy("26/1/2012"), linetype="longdash",
color = "black", size=1, )
geom_vline(xintercept = dmy("16/9/19"), linetype="dashed",
color = "black", size=1, )
geom_vline(xintercept = dmy("7/11/19"), linetype="longdash",
color = "black", size=1, )
geom_hline(yintercept =0.4, linetype="dotted",
color = "black", size=1, )
annotate(y= 0.4, x = dmy("01/5/11"), label="Normal Limit", geom = "label")
This is how I'd like plot to appear, but with "Normal limit" label on left hand facet only:
This is the data:
> dput(data)
structure(list(Date = structure(c(18873, 18843, 18766, 18746,
18745, 18738, 18705, 18691, 18688, 18645, 18583, 18549, 18541,
18509, 18502, 18457, 18435, 18381, 18380, 18362, 18320, 18276,
18232, 18219, 18207, 18199, 18195, 18194, 18190, 18184, 18180,
18177, 18164, 18152, 18148, 18127, 18108, 18095, 18066, 18038,
18016, 17988, 17948, 17919, 17875, 17858, 17830, 17802, 17763,
17732, 17704, 17689, 17669, 17646, 17618, 17583, 17557, 17520,
17487, 17443, 17400, 17381, 17373, 17368, 17340, 17312, 17284,
17249, 17232, 17228, 17207, 17196, 17186, 17134, 17105, 17088,
17078, 17043, 17037, 17032, 17016, 17008, 16986, 16951, 16930,
16915, 16910, 16878, 16860, 16812, 16766, 16729, 16715, 16701,
16689, 16674, 16671, 16668, 16661, 16657, 16643, 16640, 16581,
16561, 16524, 16489, 16443, 16402, 16365, 16344, 16310, 16304,
16286, 16255, 16224, 16204, 16189, 16175, 16160, 16150, 16141,
16127, 16120, 16105, 16090, 16077, 16069, 16057, 16057, 16048,
16041, 16034, 16024, 16021, 16014, 16008, 16002, 15996, 15993,
15988, 15981, 15975, 15973, 15964, 15940, 15926, 15908, 15883,
15875, 15870, 15861, 15849, 15845, 15838, 15835, 15832, 15828,
15828, 15826, 15819, 15819, 15798, 15783, 15765, 15715, 15674,
15660, 15635, 15602, 15593, 15572, 15552, 15517, 15489, 15478,
15455, 15427, 15399, 15380, 15365, 15364, 15362, 15355, NA, 15308,
15273, 15260, 15250, 15230, 15226, 15180, 15175, 15154, 15114,
15082, 15058, 15028, 14995, 14973, 14935, 14911, 14879, 14837,
14816, 14790, 14757, 14727, 14697, 14666, 14607, 14580, 14536,
14517, 14480, 14452, 14391, 14363, 14328, 14295, 14272, 14207,
14174, 14146, 14118, 14081, 14055, 14022, 13992, 13845, 13817,
13782, 13719, 13691, 13661), class = "Date"), Result = c(0.1,
0.1, 0.2, 0.1, 0.1, 0.2, 0.3, 0.1, 0.1, 0.2, 0.1, 0.2, 0.2, 0.2,
0.2, 0.2, 0.2, 0.2, 0.2, 0.3, 0.4, 1, 1, 1.2, 1.3, 0.9, 1.1,
1.2, 1, 0.8, 0.6, 0.5, 0.2, 0.1, 0.1, 0.2, 0.2, 0.2, 0.2, 0.1,
0.2, 0.1, 0.2, 0.1, 0.2, 0.3, 0.3, 0.3, 0.2, 0.3, 0.3, 0.2, 0.2,
0.2, 0.4, 0.3, 0.2, 0.3, 0.3, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2,
0.2, 0.2, 0.2, 0.2, 0.3, 0.3, 0.3, 0.1, 0.2, 0.2, 0.2, 0.2, 0.2,
0.2, 0.1, 0.2, 0.2, 0.2, 0.2, 0.3, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2,
0.2, 0.3, 0.2, 0.2, 0.2, 0.2, 0.2, 0.3, 0.2, 0.2, 0.4, 0.4, 0.3,
0.3, 0.3, 0.3, 0.3, 0.33, 0.29, 0.25, 0.25, 0.29, 0.36, 0.16,
0.18, 0.36, 0.29, 0.26, 0.24, 0.25, 0.33, 0.22, 0.34, 0.39, 0.4,
0.22, 0.3, 0.29, 0.31, 0.29, 0.37, 0.85, 0.52, 0.29, 0.37, 0.4,
0.4, 0.43, 0.4, 0.32, 0.27, 0.18, 0.27, 0.2, 0.18, 0.22, 0.26,
0.28, 0.27, 0.18, 0.18, 0.21, 0.21, 0.23, 0.16, 0.21, 0.21, 0.2,
0.28, 0.39, 0.27, 0.34, 0.37, 0.31, 0.33, 0.42, 0.25, 0.31, 0.31,
0.23, 0.31, 0.33, 0.4, 0.47, 0.46, 1.19, 2.51, 4.62, 5.57, 2.8,
6.05, NA, 1.21, 0.56, 0.21, 0.34, 0.72, 0.72, 0.24, 0.28, 0.25,
0.2, 0.2, 0.3, 0.22, 0.29, 0.4, 0.2, 0.2, 0.3, 0.3, 0.3, 0.2,
0.2, 0.2, 0.3, 0.2, 0.3, 0.2, 0.2, 0.2, 0.3, 0.3, 0.3, 0.2, 0.2,
0.3, 0.2, 0.2, 0.3, 0.3, 0.2, 0.3, 0.3, 0.2, 0.2, 0.2, 0.2, 0.3,
0.24, 0.32, 0.2)), row.names = c(NA, -234L), class = c("tbl_df",
"tbl", "data.frame"))
CodePudding user response:
The problem with facet-wise annotations is that annotate() do not take a data argument wherein facetting variables can be looked up. Instead, I suggest you use a regular geom_label() layer, wherein the data argument contains the facetting variable. Simplified example below:
library(ggplot2)
library(dplyr)
library(lubridate)
# data <- structure(...) # As per example data
# df <- data %>% filter(...) # As per example code
ggplot(df, aes(Date, Result))
geom_line()
facet_wrap(bin ~ ., scale = "free_x")
geom_label(
data = data.frame(bin = FALSE),
aes(x = dmy("01/5/11"), y = 0.4, label = "Normal\nLimits")
)
CodePudding user response:
I've tangled with this issue before and the best solution I found was to use grid graphics to overlay my annotations, e.g.
library(tidyverse)
library(lubridate)
#>
#> Attaching package: 'lubridate'
#> The following objects are masked from 'package:base':
#>
#> date, intersect, setdiff, union
library(grid)
library(gridExtra)
#>
#> Attaching package: 'gridExtra'
#> The following object is masked from 'package:dplyr':
#>
#> combine
df1 <- structure(list(Date = structure(c(18873, 18843, 18766, 18746,
18745, 18738, 18705, 18691, 18688, 18645, 18583, 18549, 18541,
18509, 18502, 18457, 18435, 18381, 18380, 18362, 18320, 18276,
18232, 18219, 18207, 18199, 18195, 18194, 18190, 18184, 18180,
18177, 18164, 18152, 18148, 18127, 18108, 18095, 18066, 18038,
18016, 17988, 17948, 17919, 17875, 17858, 17830, 17802, 17763,
17732, 17704, 17689, 17669, 17646, 17618, 17583, 17557, 17520,
17487, 17443, 17400, 17381, 17373, 17368, 17340, 17312, 17284,
17249, 17232, 17228, 17207, 17196, 17186, 17134, 17105, 17088,
17078, 17043, 17037, 17032, 17016, 17008, 16986, 16951, 16930,
16915, 16910, 16878, 16860, 16812, 16766, 16729, 16715, 16701,
16689, 16674, 16671, 16668, 16661, 16657, 16643, 16640, 16581,
16561, 16524, 16489, 16443, 16402, 16365, 16344, 16310, 16304,
16286, 16255, 16224, 16204, 16189, 16175, 16160, 16150, 16141,
16127, 16120, 16105, 16090, 16077, 16069, 16057, 16057, 16048,
16041, 16034, 16024, 16021, 16014, 16008, 16002, 15996, 15993,
15988, 15981, 15975, 15973, 15964, 15940, 15926, 15908, 15883,
15875, 15870, 15861, 15849, 15845, 15838, 15835, 15832, 15828,
15828, 15826, 15819, 15819, 15798, 15783, 15765, 15715, 15674,
15660, 15635, 15602, 15593, 15572, 15552, 15517, 15489, 15478,
15455, 15427, 15399, 15380, 15365, 15364, 15362, 15355, NA, 15308,
15273, 15260, 15250, 15230, 15226, 15180, 15175, 15154, 15114,
15082, 15058, 15028, 14995, 14973, 14935, 14911, 14879, 14837,
14816, 14790, 14757, 14727, 14697, 14666, 14607, 14580, 14536,
14517, 14480, 14452, 14391, 14363, 14328, 14295, 14272, 14207,
14174, 14146, 14118, 14081, 14055, 14022, 13992, 13845, 13817,
13782, 13719, 13691, 13661), class = "Date"), Result = c(0.1,
0.1, 0.2, 0.1, 0.1, 0.2, 0.3, 0.1, 0.1, 0.2, 0.1, 0.2, 0.2, 0.2,
0.2, 0.2, 0.2, 0.2, 0.2, 0.3, 0.4, 1, 1, 1.2, 1.3, 0.9, 1.1,
1.2, 1, 0.8, 0.6, 0.5, 0.2, 0.1, 0.1, 0.2, 0.2, 0.2, 0.2, 0.1,
0.2, 0.1, 0.2, 0.1, 0.2, 0.3, 0.3, 0.3, 0.2, 0.3, 0.3, 0.2, 0.2,
0.2, 0.4, 0.3, 0.2, 0.3, 0.3, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2,
0.2, 0.2, 0.2, 0.2, 0.3, 0.3, 0.3, 0.1, 0.2, 0.2, 0.2, 0.2, 0.2,
0.2, 0.1, 0.2, 0.2, 0.2, 0.2, 0.3, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2,
0.2, 0.3, 0.2, 0.2, 0.2, 0.2, 0.2, 0.3, 0.2, 0.2, 0.4, 0.4, 0.3,
0.3, 0.3, 0.3, 0.3, 0.33, 0.29, 0.25, 0.25, 0.29, 0.36, 0.16,
0.18, 0.36, 0.29, 0.26, 0.24, 0.25, 0.33, 0.22, 0.34, 0.39, 0.4,
0.22, 0.3, 0.29, 0.31, 0.29, 0.37, 0.85, 0.52, 0.29, 0.37, 0.4,
0.4, 0.43, 0.4, 0.32, 0.27, 0.18, 0.27, 0.2, 0.18, 0.22, 0.26,
0.28, 0.27, 0.18, 0.18, 0.21, 0.21, 0.23, 0.16, 0.21, 0.21, 0.2,
0.28, 0.39, 0.27, 0.34, 0.37, 0.31, 0.33, 0.42, 0.25, 0.31, 0.31,
0.23, 0.31, 0.33, 0.4, 0.47, 0.46, 1.19, 2.51, 4.62, 5.57, 2.8,
6.05, NA, 1.21, 0.56, 0.21, 0.34, 0.72, 0.72, 0.24, 0.28, 0.25,
0.2, 0.2, 0.3, 0.22, 0.29, 0.4, 0.2, 0.2, 0.3, 0.3, 0.3, 0.2,
0.2, 0.2, 0.3, 0.2, 0.3, 0.2, 0.2, 0.2, 0.3, 0.3, 0.3, 0.2, 0.2,
0.3, 0.2, 0.2, 0.3, 0.3, 0.2, 0.3, 0.3, 0.2, 0.2, 0.2, 0.2, 0.3,
0.24, 0.32, 0.2)), row.names = c(NA, -234L), class = c("tbl_df",
"tbl", "data.frame"))
df <- df1 %>%
filter((Date >= dmy("25/03/2011") & Date <= dmy("18/05/2012")) | #filter unwanted data
(Date >= dmy("18/07/2019") & Date <= dmy("28/04/2020") ))%>%
mutate(bin = ifelse(Date >= dmy("18/07/2019"), "yes", "no"))
plot = ggplot(df, aes(Date, Result))
geom_point(colour = "red3", size = 2.5)
geom_line( colour= "purple4", size = 1)
facet_wrap(bin ~ ., scale = 'free_x')
scale_x_date(date_labels = "%b %Y",guide = guide_axis(angle = 45), date_breaks = "2 months")
scale_y_continuous(breaks = seq(0,6.2, by=0.5))
theme(strip.text.x = element_blank()) # this removes the true/false bin identification
geom_vline(xintercept = dmy("29/8/2011"), linetype="dashed",
color = "black", size=1)
geom_vline(xintercept = dmy("26/1/2012"), linetype="longdash",
color = "black", size=1)
geom_vline(xintercept = dmy("16/9/19"), linetype="dashed",
color = "black", size=1)
geom_vline(xintercept = dmy("7/11/19"), linetype="longdash",
color = "black", size=1)
geom_hline(yintercept =0.4, linetype="dotted",
color = "black", size=1)
# annotate(y= 0.4, x = dmy("01/5/11"), label="Normal Limit", geom = "label")
gtext <- grid::textGrob("Normal Limit", x = unit(0.15, "npc"), y = unit(0.25, "npc"))
gcombined <- grid::grobTree(ggplotGrob(plot), gtext)
grid.arrange(gcombined)
Created on 2021-09-12 by the reprex package (v2.0.1)
Would this be suitable for your problem?