I'm using the weightloss dataset:
structure(list(id = structure(c(1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L,
9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L,
12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L,
3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L), .Label = c("1", "2",
"3", "4", "5", "6", "7", "8", "9", "10", "11", "12"), class = "factor"),
diet = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("no", "yes"), class = "factor"),
exercises = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("no", "yes"
), class = "factor"), t1 = c(10.43, 11.59, 11.35, 11.12,
9.5, 9.5, 11.12, 12.51, 11.35, 11.12, 11.12, 10.2, 11.12,
9.96, 12.05, 8.11, 12.05, 12.05, 12.28, 10.66, 11.35, 10.2,
10.2, 9.5, 10.2, 12.98, 13.21, 10.2, 11.59, 12.05, 11.59,
12.05, 11.82, 11.12, 12.51, 11.59, 10.43, 11.35, 11.82, 10.2,
13.67, 10.66, 10.2, 12.05, 11.82, 10.43, 12.74, 11.35), t2 = c(13.21,
10.66, 11.12, 9.5, 9.73, 12.74, 12.51, 12.28, 11.59, 10.66,
13.44, 11.35, 12.51, 12.74, 13.67, 14.37, 14.6, 12.98, 12.05,
14.37, 14.6, 11.82, 14.13, 13.21, 12.51, 12.98, 11.12, 9.73,
13.44, 13.67, 12.98, 11.35, 12.05, 15.29, 11.82, 12.05, 12.51,
14.83, 13.9, 13.21, 14.13, 15.06, 12.98, 11.35, 12.51, 14.13,
12.74, 11.35), t3 = c(11.59, 13.21, 11.35, 11.12, 12.28,
10.43, 11.59, 12.74, 9.96, 11.35, 10.66, 11.12, 15.76, 16.68,
17.84, 14.6, 17.84, 17.61, 18.54, 16.91, 15.52, 17.38, 19,
14.13, 14.6, 14.6, 12.05, 15.52, 13.9, 12.74, 13.21, 14.83,
14.6, 10.89, 15.52, 12.98, 14.37, 15.06, 13.44, 14.13, 15.29,
14.6, 15.06, 15.52, 13.9, 14.37, 15.06, 15.06)), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -48L))
So far it looks like at least here I can separate the scores:
weight <- weightloss
summary <- weight %>%
get_summary_stats(type = "mean_sd")
summary
Which gives me this:
A tibble: 3 x 4
variable n mean sd
<chr> <dbl> <dbl> <dbl>
1 t1 48 11.2 1.09
2 t2 48 12.7 1.42
3 t3 48 14.2 2.26
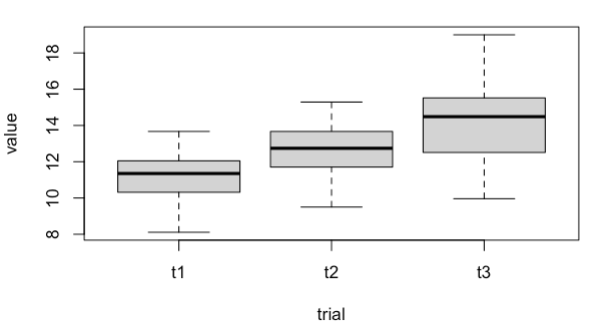
I'm trying to run a RMANOVA on this, but I would like to get a boxplot for every one of the three groups, all in a single plot. However, I'm not sure how to plot the x and y in this case. I tried using this for the x:
trial_type <- c("t1","t2","t3")
factor(trial_type)
But thats where I'm stuck...I'm not sure how you get the y in this case. The y is clearly the scores from each trial. I tried grouping by this factor to see if that would sort out the scores in some way, but I haven't figured that out either.
I'm just not sure how you plot this into ggplot. Any help would be great! I can imagine this is a very useful skill to learn for any data that uses trials.
CodePudding user response:
You may have to pivot_longer first, then the grouping gets easier.
After pivoting, all values will be on the same column ('values'), and there will be a grouping column ('trial')
library(dplyr)
df<-df %>% pivot_longer(names_to = 'trial', values_to = 'value', cols = matches('t\\d'))
with(df, boxplot(value ~ trial))
If you prefer ggplot:
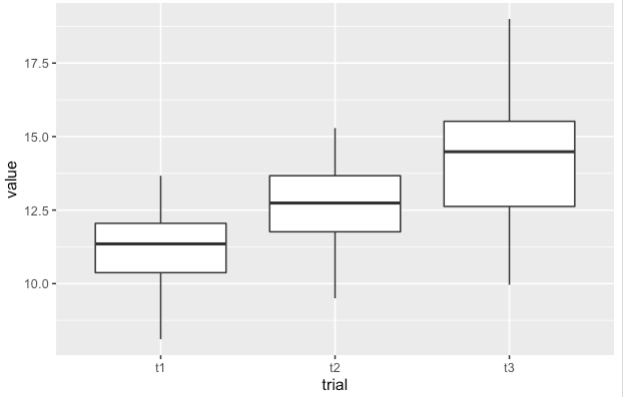
ggplot(df, aes(x=trial, y=value))
geom_boxplot()
CodePudding user response:
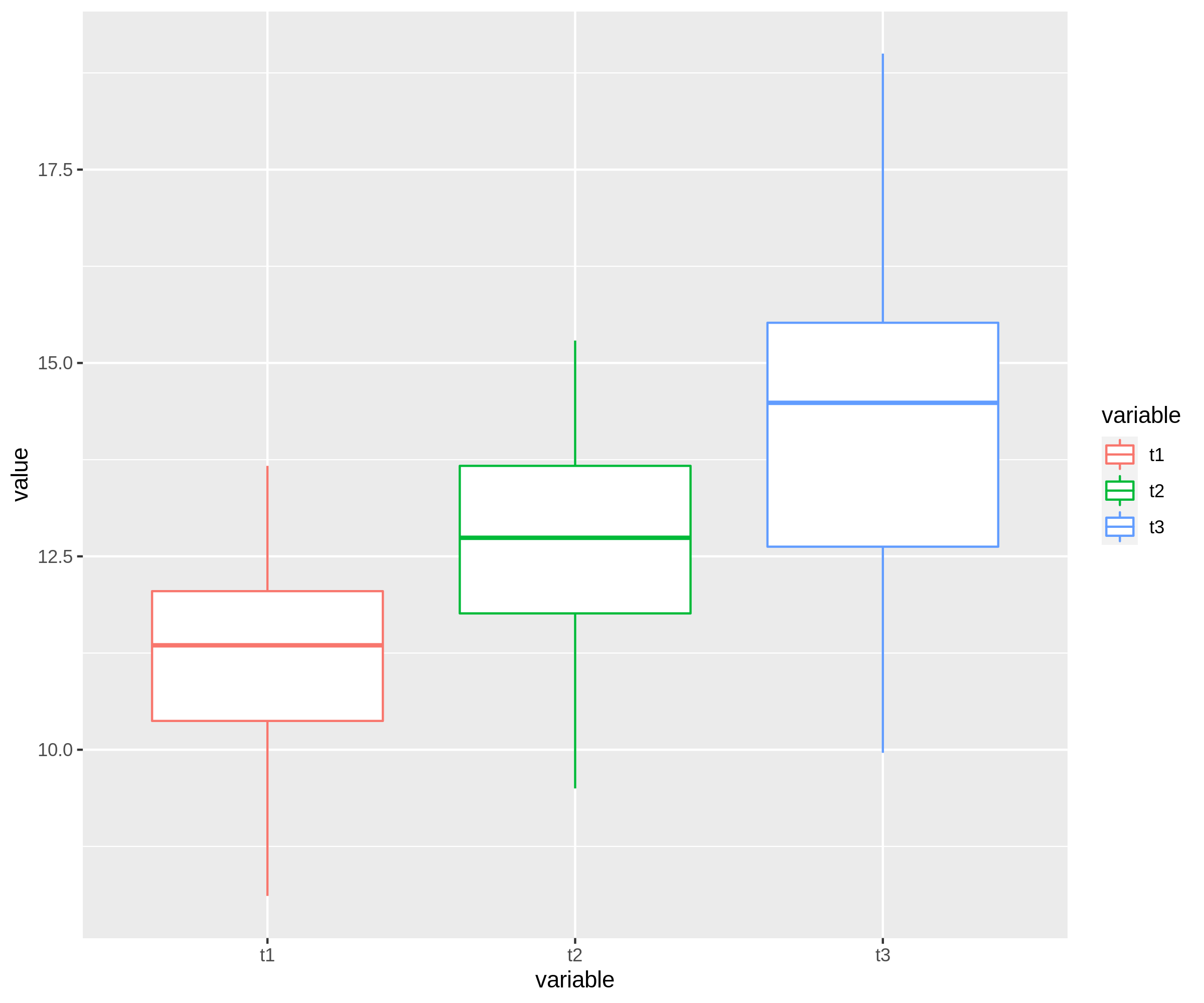
a ggplot and reshape2 way;
library(reshape2)
df %>%
melt(id.vars='id',measure.vars = c('t1','t2','t3')) %>%
ggplot(aes(x=variable,y=value))
geom_boxplot(aes(color=variable))