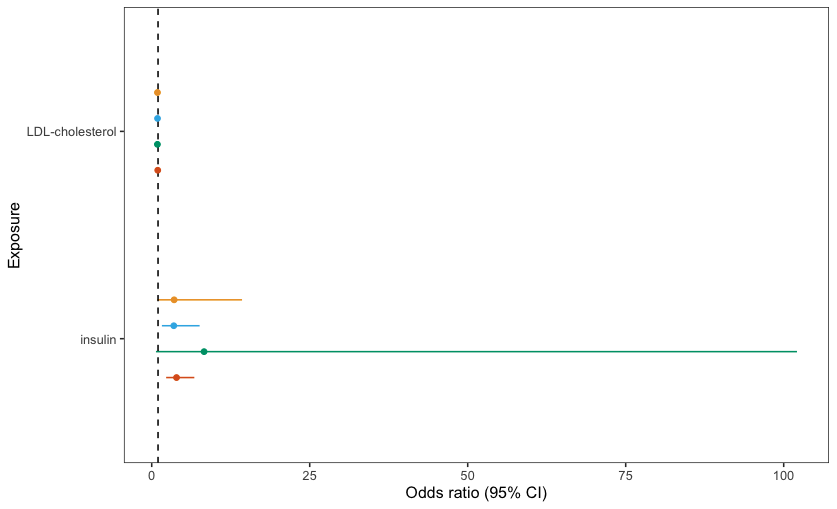
I have produced the following graph:
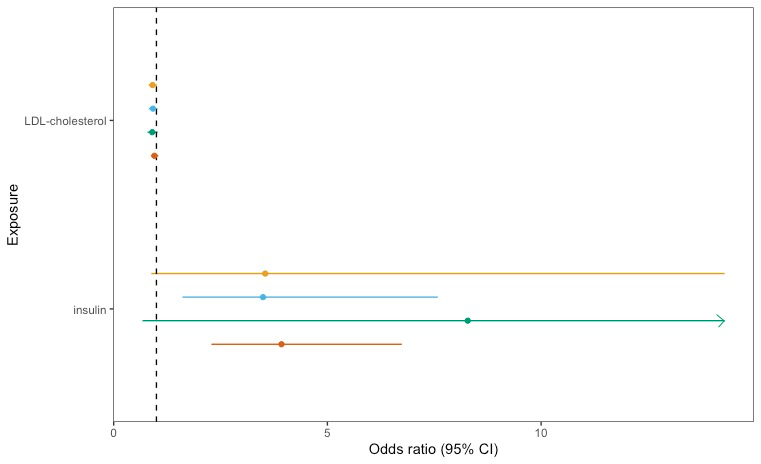
As you can see, due to one long error bar the results aren't very clear. Therefore, I want to cut-off the long error bar and replace with an arrow, like this:
However, I can't get the arrow in the right place using ggplot. Based on similar questions I have produced the following code:
p <- ggplot(dat, aes(x=exposure, y=OR, ymin=LCI, ymax=UCI,col=method))
geom_linerange(position=position_dodge(width = 0.5))
geom_hline(yintercept=1, lty=2)
geom_point(stroke = 0.5,position=position_dodge(width = 0.5))
scale_x_discrete(name="Exposure")
scale_y_continuous(name="Odds ratio (95% CI)")
coord_flip()
scale_colour_manual(values=c("#D55E00","#009E73", "#56B4E9","#E69F00"))
theme_bw() theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),text = element_text(size=11),legend.position="none")
geom_segment(aes(x = 1, y = 14, xend = 1, yend = 14), size=0.25,
arrow = arrow(length = unit(0.13, "cm")))
However, this produces the following graph:
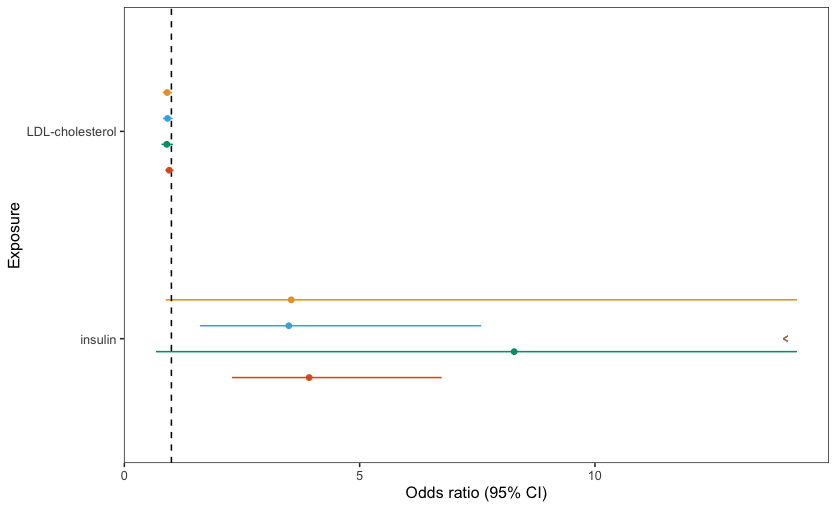
How do I make it so there is only one arrow which is in the right place (at the end of the green line and rotated 180')?
Thanks!
My data looks like:
dat <-
structure(list(outcome = c("cancer", "cancer", "cancer", "cancer",
"cancer", "cancer", "cancer", "cancer"), exposure = c("insulin",
"insulin", "insulin", "insulin", "LDL-cholesterol", "LDL-cholesterol",
"LDL-cholesterol", "LDL-cholesterol"), OR = c(3.925713203, 3.49400025,
3.545796066, 8.283204105, 0.910187777, 0.902219745, 0.918412256,
0.953392987), LCI = c(2.285899133, 1.609867994, 0.879681108,
0.672007727, 0.817076527, 0.789469504, 0.82104523, 0.870226921
), UCI = c(6.741865347, 7.583253904, 14.29230392, 14.292304,
1.013909666, 1.03107272, 1.027325952, 1.044507088), method = c("Inverse variance weighted",
"Weighted median", "Weighted mode", "MR Egger", "Weighted mode",
"MR Egger", "Weighted median", "Inverse variance weighted")), row.names = c(73L,
74L, 75L, 76L, 139L, 140L, 141L, 142L), class = "data.frame")
CodePudding user response:
The answer is actually very simple - just change the "x = 1, y = 14, xend = 1, yend = 14" within geom_segment(aes()) to place the arrow in the right place, and make the y very slightly smaller than yend to make the arrow face the right way.