I came across a rather puzzling result: pandas at assignment is almost 10 times slower than appending a list and converting it to dataframe. Why is this? I understand that pandas at deals with a little more complicated structure, but 10 times slower performance seems extreme, rendering thereby clumsy list appending more efficient. I see many posts about slowness of loc but loc does a lot more than at so in a way one can accept its sluggishness.
import pandas as pd
import numpy as np
import time
dt = pd.date_range('2010-01-01','2021-01-01', freq='H')
s = pd.Series(data = np.random.randint(0,1000, len(dt)), index = np.random.choice(dt, size=len(dt), replace=False))
d = pd.Series(data = np.nan, index=dt)
t = time.time()
for i in s.iteritems():
d.at[i[0]] = np.random.randint(10)
print(time.time()-t)
m = []
t = time.time()
for i in s.iteritems():
m.append((i[0], np.random.randint(10)))
m = pd.DataFrame(m)
m.columns = ['date_time', 'data']
m.set_index('date_time', inplace=True)
print(time.time()-t)
output:
4.053529500961304
0.3882014751434326
CodePudding user response:
Well... There is no need to use at here... Pandas isn't meant to be fast...
The append method automatically appends to the end of the list... Where at doesn't... Also you're using indexed assignment... Base Python surely would be faster.
The fastest would be bare numpy here:
import pandas as pd
import numpy as np
import time
dt = pd.date_range('2010-01-01','2021-01-01', freq='H')
s = pd.Series(data = np.random.randint(0,1000, len(dt)), index = np.random.choice(dt, size=len(dt), replace=False))
d = pd.Series(data = np.nan, index=dt)
t = time.time()
d[:] = np.random.randint(0, 10, len(dt))
print(d)
print(time.time()-t)
m = []
t = time.time()
for i in s.iteritems():
m.append((i[0], np.random.randint(10)))
m = pd.DataFrame(m)
m.columns = ['date_time', 'load']
m.set_index('date_time', inplace=True)
print(m)
print(time.time()-t)
Output:
2010-01-01 00:00:00 5
2010-01-01 01:00:00 5
2010-01-01 02:00:00 6
2010-01-01 03:00:00 6
2010-01-01 04:00:00 8
..
2020-12-31 20:00:00 7
2020-12-31 21:00:00 6
2020-12-31 22:00:00 4
2020-12-31 23:00:00 2
2021-01-01 00:00:00 1
Freq: H, Length: 96433, dtype: int32
0.03117227554321289
load
date_time
2018-04-14 04:00:00 6
2014-03-03 23:00:00 2
2010-03-11 20:00:00 1
2017-06-27 16:00:00 2
2020-11-08 11:00:00 6
... ...
2016-12-03 08:00:00 9
2020-01-12 16:00:00 5
2014-08-08 10:00:00 5
2012-05-23 04:00:00 8
2010-11-05 06:00:00 2
[96433 rows x 1 columns]
0.4729738235473633
As you can see, NumPy is like 10 times faster already for this minor dataset. NumPy is developed some part in C, and it's super quick. Just printed it out to prove that the outputs are the same.
CodePudding user response:
pandas will always lose to a list for these sort of simple operations. pandas is written in Python, and executes a ton of code at the Python interpreter level. In contrast, list.append, is written entirely in C. You just have to resolve the method and you are into the C layer.
Both operations are Constant Time
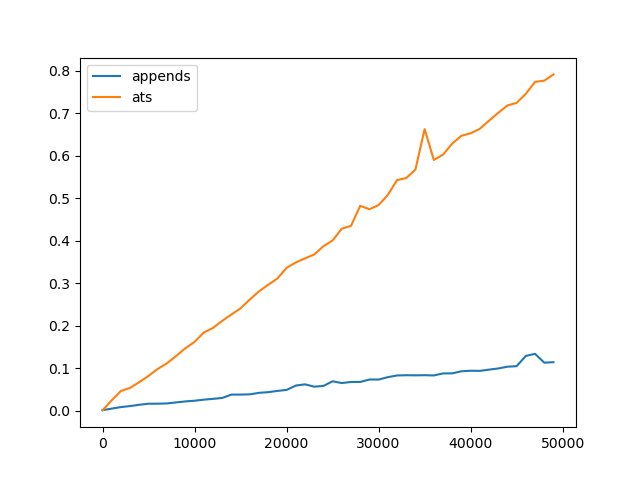
Note, both operations are constant time, but the constant factors for pandas.DataFrame.at.__setitem__ are much higher than those for list.append. Consider the following timings:
from timeit import timeit
import pandas as pd, matplotlib.pyplot as plt
setup = """
import pandas as pd
d = pd.Series(float('nan'), index=range({}))
def use_append(s):
m = []
for i,v in s.iteritems():
m.append((i, 42))
return pd.DataFrame(m)
def use_at(s):
for i,v in s.iteritems():
s[i] = 42
return pd.DataFrame(s)
"""
N = range(0, 50_000, 1000)
appends = [timeit("use_append(d)", setup.format(n), number=5) for n in N]
ats = [timeit("use_at(d)", setup.format(n), number=5) for n in N]
pd.DataFrame(dict(appends=appends, ats=ats), index=N).plot()
plt.savefig("list_append-vs-df_at.png")
As you can see, the overall algorithm is linear in both cases.
List append Implementation
To reiterate, list.append is much faster because it is entirely implemented in C. Here is the source code
static int
app1(PyListObject *self, PyObject *v)
{
Py_ssize_t n = PyList_GET_SIZE(self);
assert (v != NULL);
if (n == PY_SSIZE_T_MAX) {
PyErr_SetString(PyExc_OverflowError,
"cannot add more objects to list");
return -1;
}
if (list_resize(self, n 1) < 0)
return -1;
Py_INCREF(v);
PyList_SET_ITEM(self, n, v);
return 0;
}
int
PyList_Append(PyObject *op, PyObject *newitem)
{
if (PyList_Check(op) && (newitem != NULL))
return app1((PyListObject *)op, newitem);
PyErr_BadInternalCall();
return -1;
}
As you can see, there is a wrapper function, and then the main function app1, executes the classic dynamic array append, resize internal buffer if necessary, then set the item at the corresponding index of the internal array (see here for the PyListObject_SET_ITEM macro definition, which you will see, just sets an array at a given index).
Pandas `at` implementation
Alternatively, consider *just a little bit* what happens when you do:some_series.at[key] = value
First, some_series.at creates a pandas.core.indexing._AtIndexer object, then, for that _AtIndexer object. __setitem__ is called:
def __setitem__(self, key, value):
if self.ndim == 2 and not self._axes_are_unique:
# GH#33041 fall back to .loc
if not isinstance(key, tuple) or not all(is_scalar(x) for x in key):
raise ValueError("Invalid call for scalar access (setting)!")
self.obj.loc[key] = value
return
return super().__setitem__(key, value)
As we see here, it actually falls back to loc if the indices aren't unique. But suppose they are, it then makes an expensive call to super().__setitem__(key, value), taking us to _ScalarAccessIndexer.__setitem__:
def __setitem__(self, key, value):
if isinstance(key, tuple):
key = tuple(com.apply_if_callable(x, self.obj) for x in key)
else:
# scalar callable may return tuple
key = com.apply_if_callable(key, self.obj)
if not isinstance(key, tuple):
key = _tuplify(self.ndim, key)
key = list(self._convert_key(key))
if len(key) != self.ndim:
raise ValueError("Not enough indexers for scalar access (setting)!")
self.obj._set_value(*key, value=value, takeable=self._takeable)
Here, it handles a special case if the key is a callable (i.e. a function), does some basic bookeeping on the type of the key and checks the length. If all goes well, it delegates to self.obj._set_value(*key, value=value, takeable=self._takeable), yet another Python function call. For completion sake, let's look at pd.Series._set_value:
def _set_value(self, label, value, takeable: bool = False):
"""
Quickly set single value at passed label.
If label is not contained, a new object is created with the label
placed at the end of the result index.
Parameters
----------
label : object
Partial indexing with MultiIndex not allowed.
value : object
Scalar value.
takeable : interpret the index as indexers, default False
"""
if not takeable:
try:
loc = self.index.get_loc(label)
except KeyError:
# set using a non-recursive method
self.loc[label] = value
return
else:
loc = label
self._set_values(loc, value)
Again, more book keeping. Note, if we actually were to grow the pandas object, it falls back to loc...
def _set_values(self, key, value) -> None:
if isinstance(key, (Index, Series)):
key = key._values
self._mgr = self._mgr.setitem(indexer=key, value=value)
self._maybe_update_cacher()
But if the size doesn't change, it finally delegates to the block manager, the internal guts of pandas objects: self._mgr.setitem(indexer=key, value=value)... Note this isn't even done yet! But I think you get the idea...