The file "Aspirin" contains a 2 × 2 × 2 contingency table with columns defined as follows.
Column 1: V1=Observation number. [Observations 1 to 8.]
Column 2: V2=Count. [Nonnegative integer count for each cell in the Table.]
Column 3: V3=Case/Control Factor. [Factor Level 1 (Controls) and Level 2 (Cases).]
Column 4: V4=Ulcer Type Factor. [Factor Level 1 (Gastric) and Level 2 (Duodenal).]
Column 5: V5=Aspirin Use Factor. [Factor Level 1 (Non-User) and Level 2 (User).]
> aspirin
V1 V2 V3 V4 V5
1 1 62 1 1 1
2 2 39 2 1 1
3 3 53 1 2 1
4 4 49 2 2 1
5 5 6 1 1 2
6 6 25 2 1 2
7 7 8 1 2 2
8 8 8 2 2 2
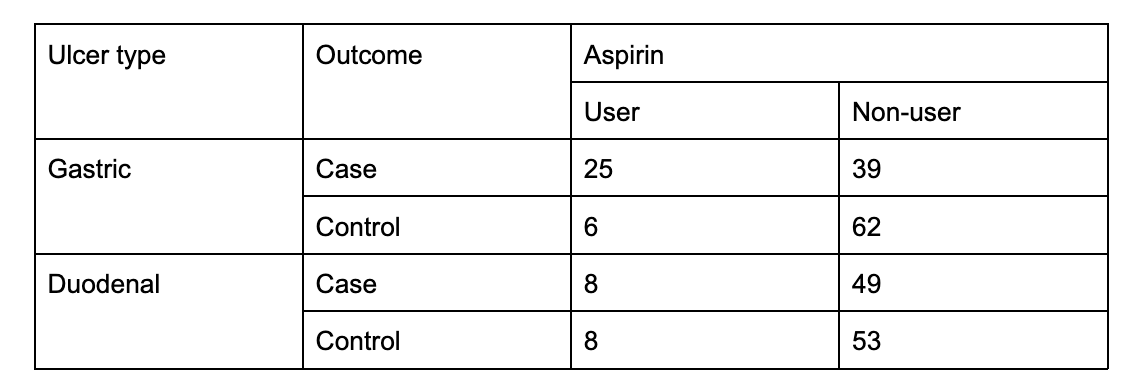 I want to construct a 2x2x2 contingency table like the image above in R, so I typed the following code:
I want to construct a 2x2x2 contingency table like the image above in R, so I typed the following code:
case_control=factor(aspirin$V3)
ulcer=factor(aspirin$V4)
use=factor(aspirin$V5)
table(case_control,ulcer,use)
But I get something like this:
, , use = 1
ulcer
case_control 1 2
1 1 1
2 1 1
, , use = 2
ulcer
case_control 1 2
1 1 1
2 1 1
I want a contingency table with counts, so obviously the result above is not what I'm desiring. Is there a way to fix this?
CodePudding user response:
In your case, just use
ftable(case_control,ulcer,use)
which returns a "flat" table
use 1 2
case_control ulcer
1 1 1 1
2 1 1
2 1 1 1
2 1 1
The main problem here is, that you are discarding your count column. So as an alternative here is a - in my opinion - better approach:
You could use xtabs together with ftable() (here used in a dplyr pipe):
library(dplyr)
df %>%
transmute(ID = V1,
Count = V2,
Case_Control = factor(V3,
labels = c("Control", "Case")),
Ulcer_Type = factor(V4,
labels = c("Gastric", "Duodenal")),
Aspirin_Use = factor(V5,
labels = c("Non-User", "User"))) %>%
xtabs(Count ~ Ulcer_Type Case_Control Aspirin_Use, data = .) %>%
ftable()
This returns
Aspirin_Use Non-User User
Ulcer_Type Case_Control
Gastric Control 62 6
Case 39 25
Duodenal Control 53 8
Case 49 8
Data
df <- structure(list(V1 = c(1, 2, 3, 4, 5, 6, 7, 8), V2 = c(62, 39,
53, 49, 6, 25, 8, 8), V3 = c(1, 2, 1, 2, 1, 2, 1, 2), V4 = c(1,
1, 2, 2, 1, 1, 2, 2), V5 = c(1, 1, 1, 1, 2, 2, 2, 2)), row.names = c(NA,
-8L), class = c("tbl_df", "tbl", "data.frame"))
