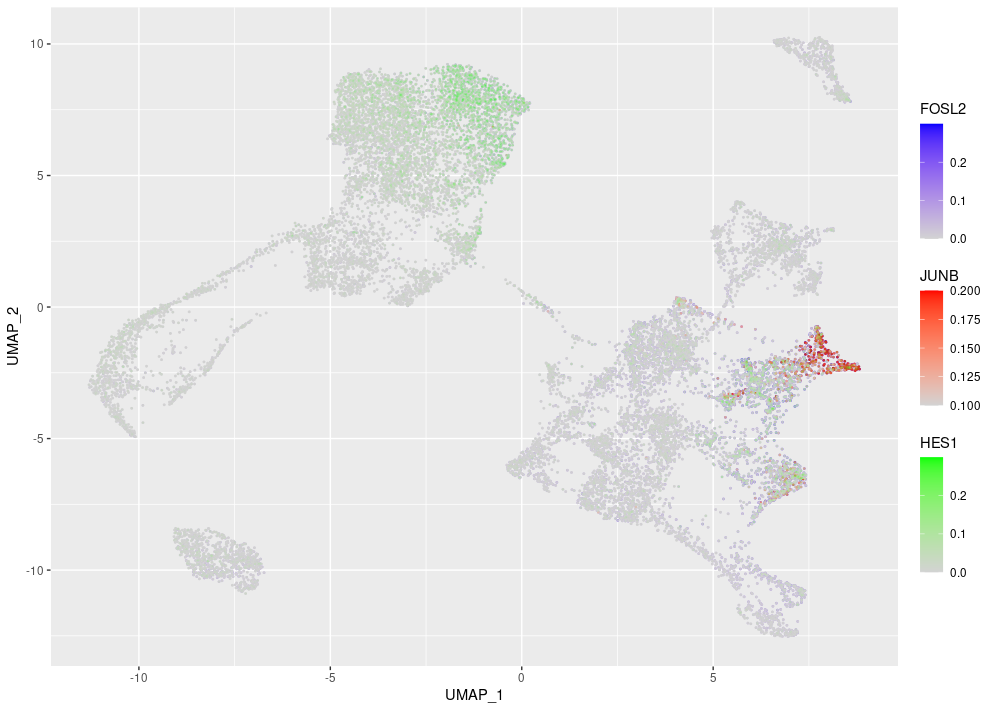
I was just wondering if anybody had any experience with coloring something like a UMAP made in ggplot based on the expression of multiple genes at the same time? What I want to do is something like the blend function in Seurat featureplots, but with 3 genes / colors instead of 2.
I'm looking to make something like this:
Where the colors for the genes combine where there is overlap.
What I've gotten to so far is
ggplot(FD, vars = c("UMAP_1", "UMAP_2", "FOSL2", "JUNB", "HES1"), aes(x = UMAP_1, y = UMAP_2, colour = FOSL2))
geom_point(size=0.3, alpha=1)
scale_colour_gradientn(colours = c("lightgrey", colour1), limits = c(0, 0.3), oob = scales::squish)
new_scale_color()
geom_point(aes(colour = JUNB), size=0.3, alpha=0.7)
scale_colour_gradientn(colours = c("lightgrey", colour2), limits = c(0.1, 0.2), oob = scales::squish)
new_scale_color()
geom_point(aes(colour = HES1), size=0.3, alpha=0.1)
scale_colour_gradientn(colours = c("lightgrey", colour3), limits = c(0, 0.3), oob = scales::squish)
Where FD is a data frame containing the information from the seurat object for the UMAP coordinates and the expression levels of the three genes of interest. All I can get is a plot where the points from one layer obscure those below it, I've tried messing around with the colours, gradients, alpha and scales but I'm guessing I'm doing it the wrong way.
If anyone knows of a way to make this work or has any suggestions on something else to try that would be very much appreciated.
CodePudding user response:
There is no 'vanilla' way of doing this in ggplot2. One can precalculate the blended colours and append invisible layers and scales with the ggnewscale package.
Let's pretend for reproducibility purposes that we want to make a UMAP of the iris dataset and using the descriptors of leaves as 'genes'.
library(ggplot2)
library(scales)
library(ggnewscale)
#> Warning: package 'ggnewscale' was built under R version 4.1.1
# Calculate a UMAP
umap <- uwot::umap(iris[, 1:4])
# Combine with original data and blended colours
df <- cbind.data.frame(
setNames(as.data.frame(umap), c("x", "y")),
iris,
colour = rgb(
rescale(iris$Sepal.Length),
rescale(iris$Sepal.Width),
rescale(iris$Petal.Length)
)
)
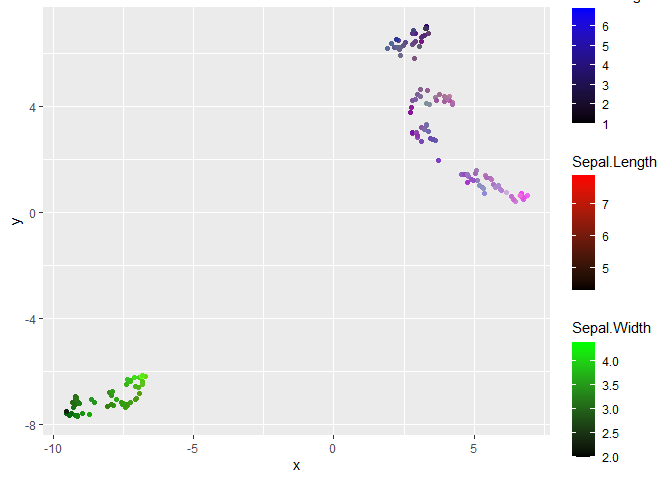
ggplot(df, aes(x, y, colour = colour))
geom_point()
scale_colour_identity()
new_scale_colour()
# shape = NA --> invisible layers
geom_point(aes(colour = Sepal.Length), shape = NA)
scale_colour_gradient(low = "black", high = "red")
new_scale_colour()
geom_point(aes(colour = Sepal.Width), shape = NA)
scale_colour_gradient(low = "black", high = "green")
new_scale_colour()
geom_point(aes(colour = Petal.Length), shape = NA)
scale_colour_gradient(low = "black", high = "blue")
#> Warning: Removed 150 rows containing missing values (geom_point).
#> Warning: Removed 150 rows containing missing values (geom_point).
#> Warning: Removed 150 rows containing missing values (geom_point).
On the more experimental side of things, I have a package on github that has related functionality.
library(ggchromatic) # devtools::install_github("teunbrand/ggchromatic")
ggplot(df, aes(x, y, colour = rgb_spec(Sepal.Length, Sepal.Width, Petal.Length)))
geom_point()
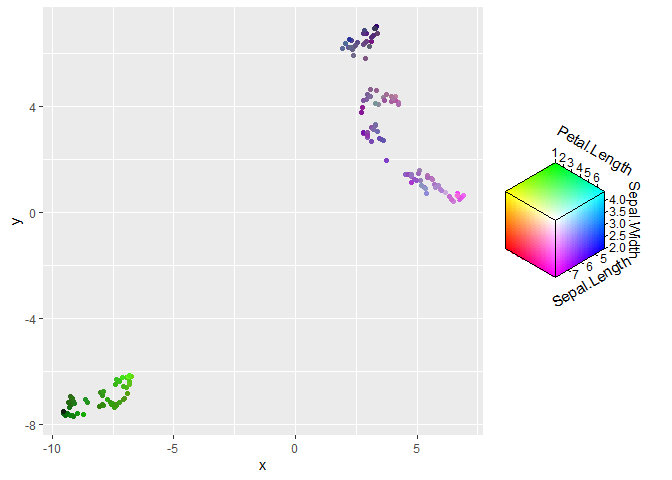
Created on 2021-10-18 by the reprex package (v2.0.1)
A small sidenote: a plot becomes very hard to interpret when some attributes of the data are mapped to different colour channels.