I need to get graphs that look consistent in ggplot2. The script is returning about 500 graphs per run, so changing it manually is not an option.
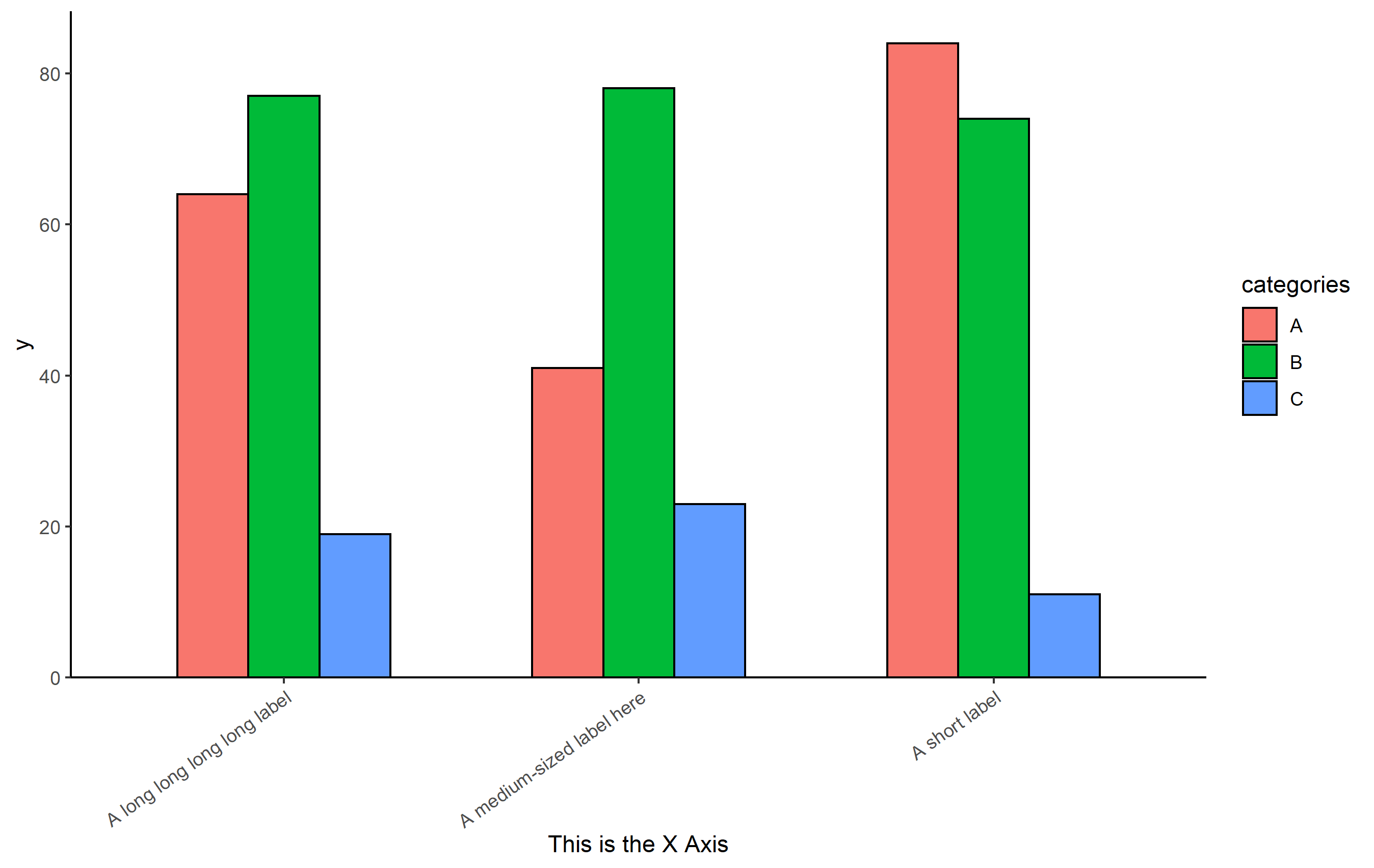
My two problems are that first, when the label is too long, the graph gets smaller (shown in Figure A) and second, when I have more conditions (more bars), the bars get narrower (shown in Figure B). I am a beginner so I need your advice as I cannot find anything online. Also, the script is not mine. It is from an ex-coworker and I am trying to improve it. It is 300 lines long so I don´t think I can post it in here.
CodePudding user response:
OP. It's a bit tough to help with your particular case without code, but here's a suggestion on how you may maintain the height of the plot to be consistent, given some variation in names on the x axis.
For the part of your question about narrowing the columns when you have more conditions... that's just going to happen. What would you like to happen as an alternative? If you can specify that, we can help - maybe put it as a separate question with a reproducible example.
Here's a reproducible example:
library(ggplot2)
set.seed(8675309)
df <- data.frame(
x = rep(c("A short label", "A long long long long label", "A medium-sized label here"), each=3),
categories = rep(LETTERS[1:3], 3),
y = sample(1:100, 9)
)
p <-
ggplot(df, aes(x=x, y=y, fill=categories))
geom_col(position=position_dodge(0.6), width=0.6, color='black')
scale_y_continuous(expand=expansion(mult=c(0, 0.05)))
xlab("This is the X Axis")
theme_classic()
theme(
axis.text.x = element_text(angle=35, hjust=1, vjust=1)
)
p
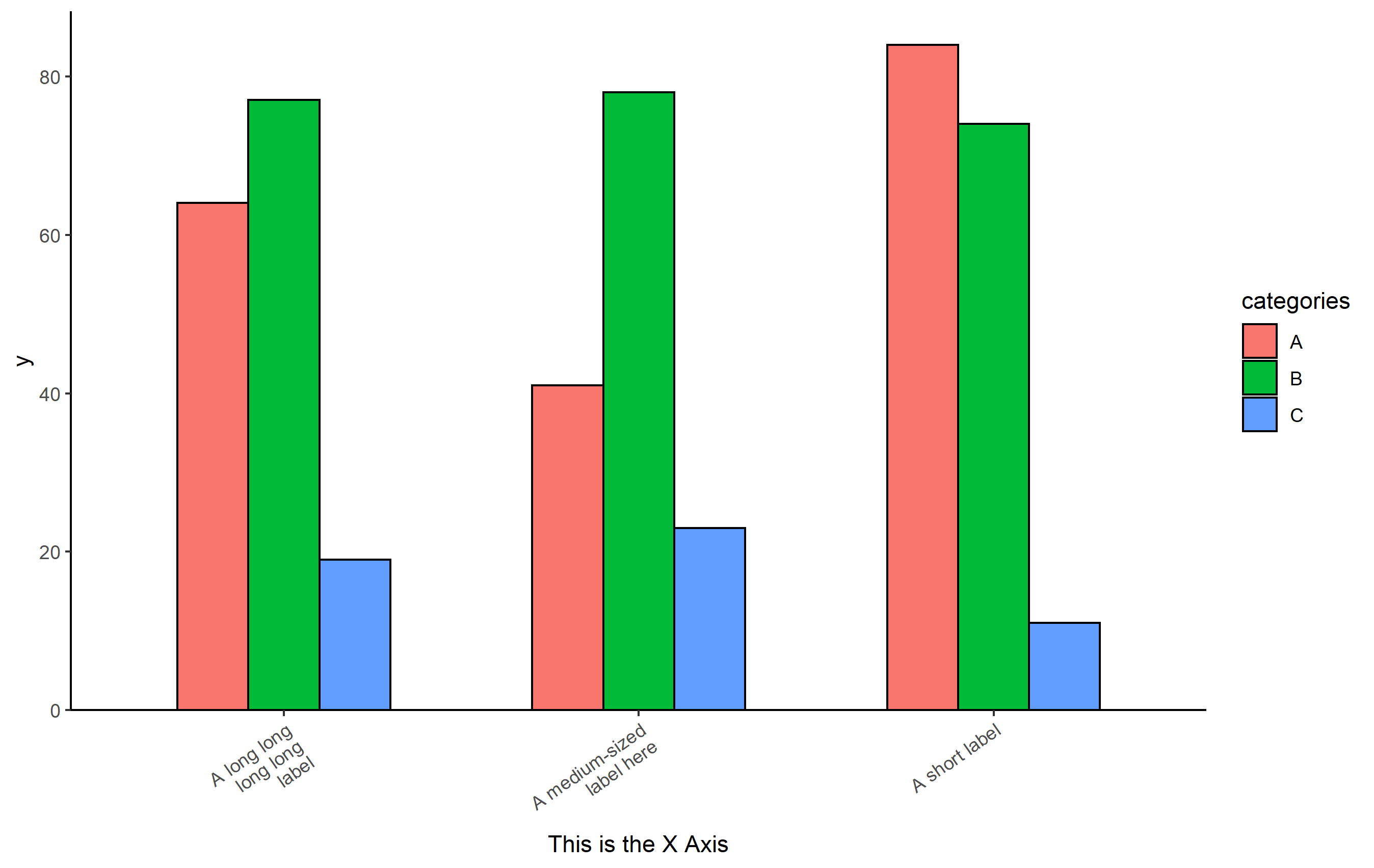
Based on that code, the distance from the bottom of the bars on the plot to the axis title will vary depending on the length of the text on the axis. One approach to solve this inconsistent look is to force text to wrap around to the next line if it is longer than a specific maximum length. I'm going to do that using the scales package:
library(scales)
# force text longer than 15 characters to wrap to the next line
p scale_x_discrete(labels=label_wrap(15))
As long as you don't have charts where everything is under 15 characters on the x axis labels... that will work. You may have to play with the precise number of characters to force wrapping.
CodePudding user response:
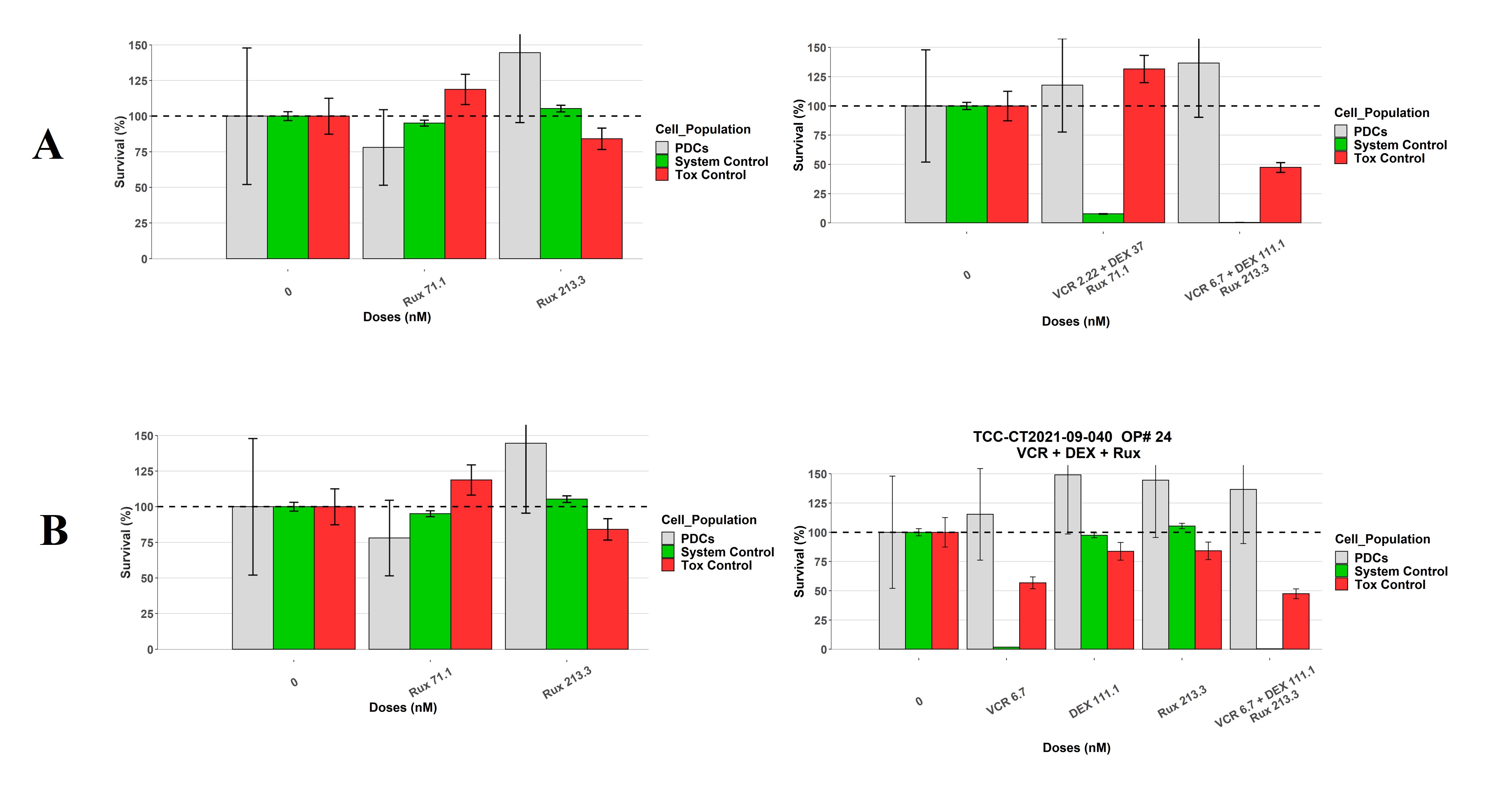
In order to narrow my columns I would like to get the size of my overall plot smaller. As shown in the next picture, when I plot 5 conditions, I get graph A (with a column size of "violet line". When I plot 4 conditions or less, I get graph B which has different column sizes as A (see that same "violet line" is now smaller than columns). I would like to get graph C, which has the same column sizes as A but it is smaller (it is just missing a column group).
I will leave the part of the script where I plot the graphs:
Blockquote
###############################################################################
# Part 1: Initialize the variables and setup the general variables (e.g. colors).
figures_case_1 <- list()
Controls_case_1 <- list()
ii <- 1
colors <- c('gray85','green3', 'firebrick1')
text_size <- 20
# Part 2: Create the table with the information to plotted.
for(l in 2:length(all_dataframes)){
splitted_doses_first <- unlist(strsplit(as.character(all_dataframes[l][[1]][1,"Dose"]),"\\s"))
# Initialize the dataframe
combined_dataFrame <- data.frame(matrix(ncol = 6, nrow = 0))
x <- c("Activity", "Dose", "STD", "Cell_Population","Drug","dose_number")
colnames(combined_dataFrame) <- x
if(length(splitted_doses_first) > 2){
# Entro a este loop cuando tengo mas de una droga. Desde dos combinaciones en adelante.
for(j in 2:length(all_dataframes)){
splitted_doses_second <- unlist(strsplit(as.character(all_dataframes[j][[1]][1,"Dose"]),"\\s"))
if(length(splitted_doses_second) == 2){
# Entro a este loop cuando tenga una droga individual.
intersection <- intersect(splitted_doses_first,splitted_doses_second)
idx <- which(intersection == ' ')
if(length(idx) != 0){intersection <- intersection[-idx]}
if(length(intersection) == 2 && str_contains(as.character(all_dataframes[l][[1]][1,"Dose"]),
as.character(all_dataframes[j][[1]][1,"Dose"]))){
aux <- all_dataframes[j][[1]]
combined_dataFrame <- rbind(aux, combined_dataFrame)
}
}
}
aux <- all_dataframes[1][[1]] # Add DMSO.
combined_dataFrame <- rbind(aux, combined_dataFrame)
# Part 3: Order the table.
combined_dataFrame$dose_number <- as.numeric(as.character(combined_dataFrame$dose_number))
combined_dataFrame <- combined_dataFrame[order(combined_dataFrame["dose_number"]),]
row.names(combined_dataFrame) <- NULL
combined_dataFrame$Dose <- factor(combined_dataFrame$Dose, levels = unique(combined_dataFrame$Dose))
combined_dataFrame <- rbind(combined_dataFrame, all_dataframes[l][[1]])
Controls_case_1[[ii]] = combined_dataFrame
# Part 4: Create the figure.
title_fig <- as.character(all_dataframes[l][[1]][1,"Drug"])
figure <- ggplot(data = combined_dataFrame, aes(x=as.factor(Dose), y=Activity, fill=Cell_Population))
geom_hline(yintercept=25, linetype="solid", colour = "grey86", size=0.5)
geom_hline(yintercept=50, linetype="solid", colour = "grey86", size=0.5)
geom_hline(yintercept=75, linetype="solid", colour = "grey86", size=0.5)
geom_hline(yintercept=125, linetype="solid", colour = "grey86", size=0.5)
geom_hline(yintercept=150, linetype="solid", colour = "grey86", size=0.5)
geom_bar(stat="identity", color="black",
width = 0.8, position = position_dodge(width = 0.9))
geom_errorbar(aes(ymin=Activity-STD, ymax=Activity STD), width=.2, alpha=0.9, size=0.5,
position=position_dodge(.9))
coord_cartesian(ylim = c(0, 150))
scale_y_continuous(breaks=c(0,25,50,75,100,125,150))
# Counts: y_axes="Events" // Normalized: y_axes = "Survival (%)" // // OnlyDMSO: x_axes= Cell_Population.
labs(title=paste(exp," OP#", patient," \n", title_fig), y = "Survival (%)", x = "Doses (nM)")
theme(
# panel.grid.major = element_line(colour = "gray48"), #LINEAS DE FONDOS
panel.background = element_rect(fill = "white"), #COLOR DE FRÁFICA FONDO # plot.margin = margin(2, 2, 2, 2, "cm"),
plot.background = element_rect(
fill = "white",
colour = "white",
size = 0.1),
plot.title = element_text(hjust = 0.5, size = text_size * 1.3, face = "bold"),
axis.text.x = element_text(size=text_size,angle=30, hjust=0.5, vjust=0.5, face = "bold"),
axis.text.y = element_text(size=text_size, face = "bold"),
axis.title = element_text(size=text_size * 1.1, face = "bold"),
legend.text= element_text(size=text_size * 1.1, face = "bold"),
legend.title = element_text(size=text_size * 1.1, face = "bold"))
geom_hline(yintercept=0, linetype="solid", colour = "black", size=0.1)
geom_segment(aes(x = 0, y = 0, xend = 0, yend = 150))
geom_hline(yintercept=100, linetype="dashed", colour = "black", size=1)
scale_fill_manual(values=colors)
# Part 5: Saving independently all the figures.
figures_case_1[[1]] = figure
g <- grid.arrange(grobs = figures_case_1, nrow = 1 ,ncol = 1,gp=gpar(fontsize=2))
ggsave(paste(ii,"-",exp,"-",norm_count," ", title_fig,'.png',sep=""), g,
device = png , path = path_CYT_tables,
width = 12, height = 6,limitsize = FALSE)
ii <- ii 1
} }