I created a test data frame, and ran a mutation on that data frame. I think this issue might occur because I used subset on the column from the count function. but I am not really sure.
df<- data.frame(treatment = rep(c("A","B","C"),times = 3),
numb = c(1:3,6:9,12,13))
df_test1 <- df %>%
group_by(treatment) %>%
summarise(mean= (mean(numb)), sum=(sum(numb))) %>%
mutate(times = count(df, treatment)[2], thing = mean sum)
write.csv(df_test1, 'test.csv')
Instead of 3 in the times column the value is c(3,3,3). Any ideas why this is happening?
CodePudding user response:
We can use n() to get the count instead of count as count expects a data.frame as input and returns a data.frame
library(dplyr)
df %>%
group_by(treatment) %>%
summarise(mean= mean(numb), sum=sum(numb), times = n()) %>%
mutate(thing = mean sum)
-output
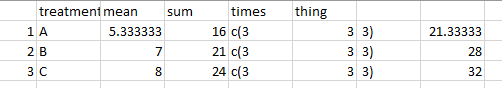
# A tibble: 3 × 5
treatment mean sum times thing
<chr> <dbl> <dbl> <int> <dbl>
1 A 5.33 16 3 21.3
2 B 7 21 3 28
3 C 8 24 3 32
If we check the structure of the output from OP's post, it becomes clear i.e. 'times' column is a data.frame as the subsetting was done as [2] instead of [[2]] to extract as a vector and [2] returns a data.frame with a single column. For data.frame, even [,2] would work as drop = TRUE by default in data.frame, whereas it wouldn't work with tibble or data.table. Safer is to use [[.
> str(df_test1)
tibble [3 × 5] (S3: tbl_df/tbl/data.frame)
$ treatment: chr [1:3] "A" "B" "C"
$ mean : num [1:3] 5.33 7 8
$ sum : num [1:3] 16 21 24
$ times :'data.frame': 3 obs. of 1 variable:
..$ n: int [1:3] 3 3 3
$ thing : num [1:3] 21.3 28 32
i.e. if we do [[2]]
df_test1 <- df %>%
group_by(treatment) %>%
summarise(mean= (mean(numb)), sum=(sum(numb))) %>%
mutate(times = count(df, treatment)[[2]], thing = mean sum)
check the structure again
> str(df_test1)
tibble [3 × 5] (S3: tbl_df/tbl/data.frame)
$ treatment: chr [1:3] "A" "B" "C"
$ mean : num [1:3] 5.33 7 8
$ sum : num [1:3] 16 21 24
$ times : int [1:3] 3 3 3
$ thing : num [1:3] 21.3 28 32