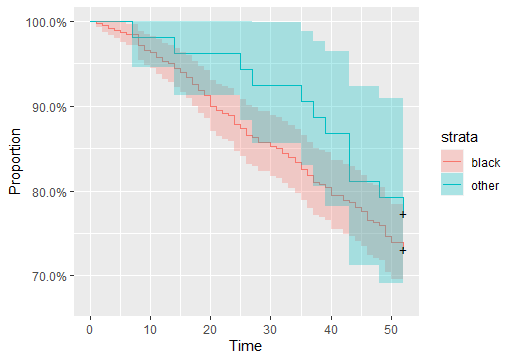
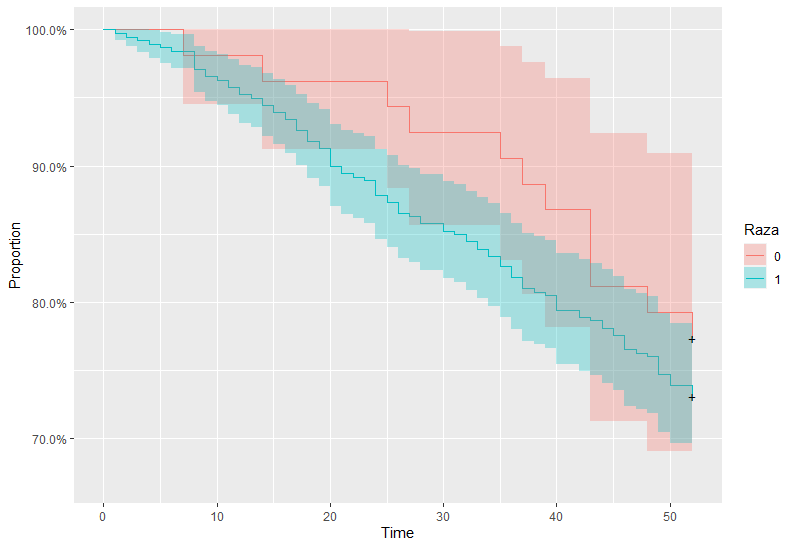
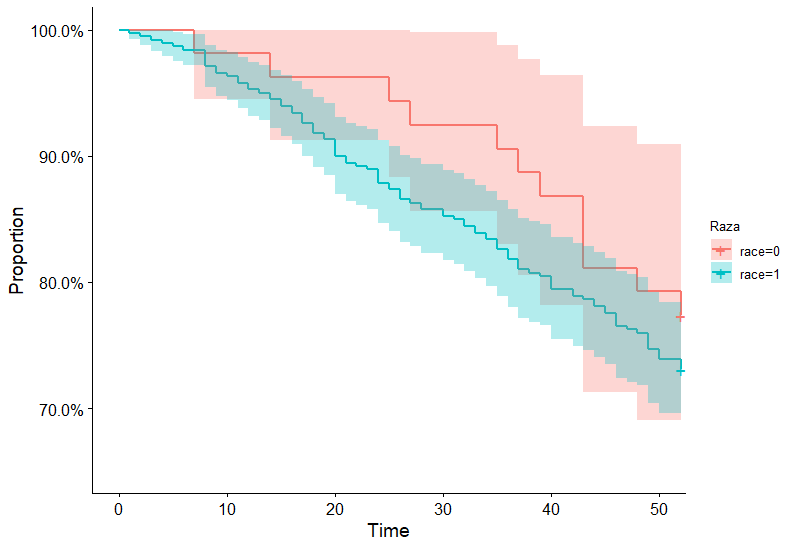
I am using Rossi data to estimate a survival curve. This is the code I used to generate the plot:
km1 <- survfit(Surv(week,arrest) ~ race,type = "kaplan-meier",data=Rossi)
autoplot(km1)
labs(x="Time", y="Proportion", strata= "Raza")
I got this result:
How can I chage the strata label to Race?
CodePudding user response:
Instead of strata = "Raza", you using both color = "Raza" and fill = "Raza" will works.
library(GlobalDeviance)
library(ggfortify)
km1 <- survfit(Surv(week,arrest) ~ race,type = "kaplan-meier",data=Rossi)
autoplot(km1)
labs(x="Time", y="Proportion", color = "Raza", fill = "Raza")
I'm not sure that colour = "Raza" works. It did't work for me.
By using survminer::ggsurvplot,
survminer::ggsurvplot(km1, xlab = "Time", ylab = "Proportion",
legend.title = "Raza", conf.int = TRUE, ylim = c(0.65,1),
legend = "right", surv.scale = "percent")