I have data such as:
in out
1 3
2 3
This represents the following tree structure:
1 2
\ /
3
Every tree will consist of sub-trees like this one, with 2 sources (in) and one destination (out). A slightly more complex tree is:
in out
1 3
2 3
3 5
4 5
which represents this tree:
1 2
\ /
3 4
\ /
5
So, the top of every tree will contain exactly 2 nodes, while the bottom will contain exactly 1 node and each level in between will contain exactly 2 nodes. There could be many levels. One final example:
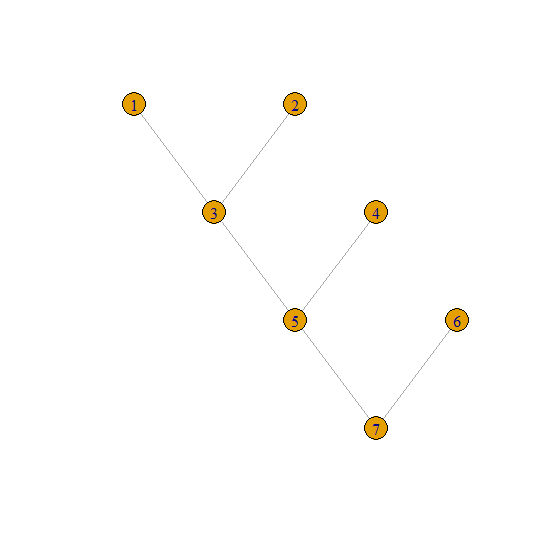
in out
1 3
2 3
3 5
4 5
5 7
6 7
which represents this tree:
1 2 level 1
\ /
3 4 level 2
\ /
5 6 level 3
\ /
7 level 4
How can I plot these trees in a nice way ? I can't use ggtree because the version of R that I have to use does not support it. I have looked at igraph and graph.tree but so far can't figure out how to do this.
If it matters, I would be happy plotting it upside down compared to how I have shown the trees above - or left to right.
CodePudding user response:
The input is an edge list so we can convert that to an igraph object and then plot it using the indicated layout. lay is a two column matrix givin gthe coordinates of the vertices and the matrix multiplication involving lay negates its second column thereby flipping it.
library(igraph)
DF <- data.frame(in. = 1:6, out. = c(3, 3, 5, 5, 7, 7)) # input
g <- graph_from_edgelist(as.matrix(DF[2:1]))
lay <- layout_as_tree(g)
plot(as.undirected(g), layout = lay %*% diag(c(1, -1)))