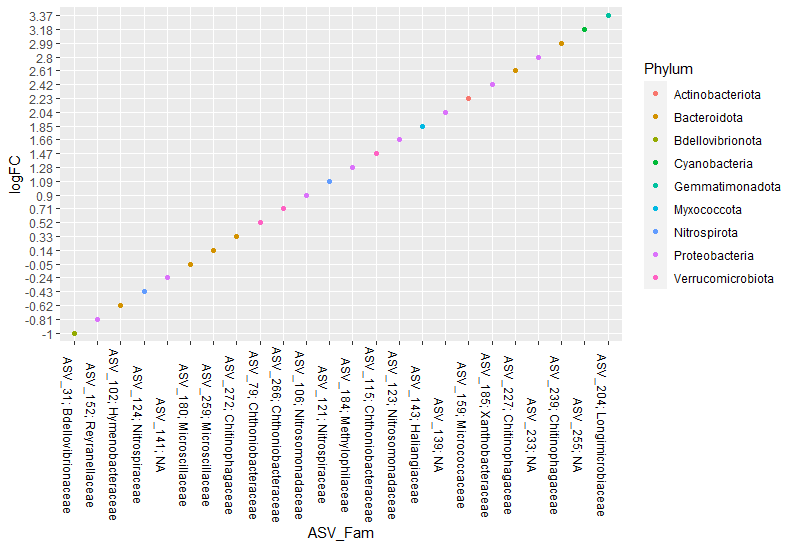
I am trying to graph using the ggplot2 geom_point call in R. However, when I plot the desired graph, my X-labels that are words (not numbers) do not all show up on the X-axis.
To begin, here is some reproducible data:
Bac <- data.frame(logFC = seq(-1, 3.5, 0.19),
ASV_Fam = c("ASV_31; Bdellovibrionaceae", "ASV_152; Reyranellaceae", "ASV_102; Hymenobacteraceae", "ASV_124; Nitrospiraceae", "ASV_141; NA",
"ASV_180; Microscillaceae", "ASV_259; Microscillaceae", "ASV_272; Chitinophagaceae", "ASV_79; Chthoniobacteraceae",
"ASV_266; Chthoniobacteraceae", "ASV_106; Nitrosomonadaceae", "ASV_121; Nitrospiraceae", "ASV_184; Methylophilaceae", "ASV_115; Chthoniobacteraceae",
"ASV_123; Nitrosomonadaceae", "ASV_143; Haliangiaceae", "ASV_139; NA", "ASV_159; Micrococcaceae", "ASV_185; Xanthobacteraceae", "ASV_227; Chitinophagaceae",
"ASV_233; NA", "ASV_239; Chitinophagaceae", "ASV_255; NA", "ASV_204; Longimicrobiaceae"),
Phylum = c("Bdellovibrionota", "Proteobacteria", "Bacteroidota", "Nitrospirota",
"Proteobacteria", "Bacteroidota", "Bacteroidota", "Bacteroidota",
"Verrucomicrobiota", "Verrucomicrobiota", "Proteobacteria", "Nitrospirota",
"Proteobacteria", "Verrucomicrobiota", "Proteobacteria", "Myxococcota",
"Proteobacteria", "Actinobacteriota", "Proteobacteria", "Bacteroidota",
"Proteobacteria", "Bacteroidota", "Cyanobacteria","Gemmatimonadota"))
Bac$Family <- gsub("^[^.]*;", "", Bac$ASV_Fam)
The closest I have found to my error is this post: 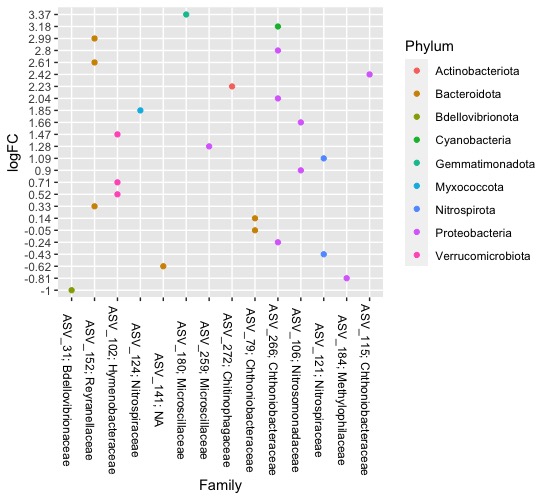
As you can see, it only plotted 14 of my 24 labels I have passed for the X-axis. All of my points are there, but some vertical lines show more than 1 point and only 1 label is associated with that vertical line. See for example X-axis labels: ASV_152; Reyranellaceae, ASV_102; Hymenobacteraceae, ASV_266; Chthoniobacteraceae, etc.
I am not sure why these are not given separate X-axis labels and are instead being graphed on the same vertical line, thus reducing the total labels plotted on the X-axis.
Other work arounds I have tried: widening the pdf via the pdf() command, widening the graph by passing coord_fixed(ratio = 0.25), but none of these options work.
In addition, passing the following code scale_y_discrete(breaks = seq(-1, 4, 0.5)) so that I can not have so many numbers show up in the Y-axis does not work. I think it's because the y-axis has been set as a factor, so I tried to keep it numeric, but that isn't working either.
Any clue into what is going on would be very helpful!
For reference, here is the ouput of my session sessionInfo()
R version 4.1.1 (2021-08-10)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur 11.6
CodePudding user response:
When you check dim(table(Bac$Family)), there exists 14 unique values in Family, and as you set x axis of your plot by x = Family, there only 14 values exists.
I'm not sure about your purpose, but if you want all 24 labels(ASV_Fam) in x axis and one point per each, instead of x = Family, try x = ASV_Fam
Please let me know if this plot is not what you wanted.
ggplot(Bac, aes(x = ASV_Fam, y = logFC, color = Phylum)) geom_point()
theme(axis.text.x = element_text(colour = "black", size = 9, angle = -90))