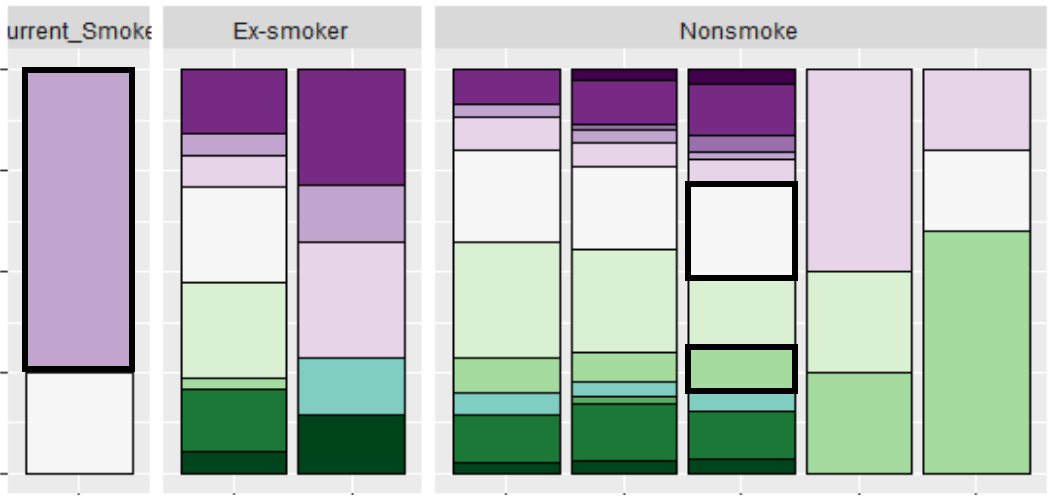
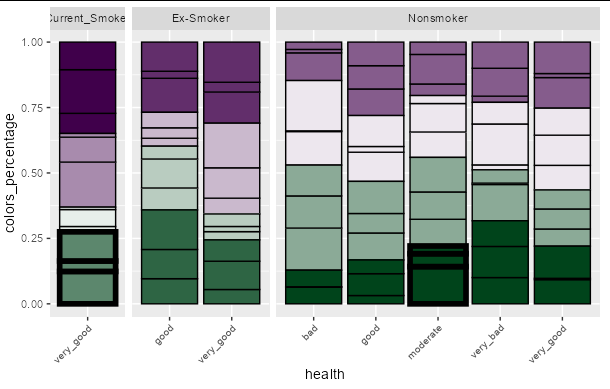
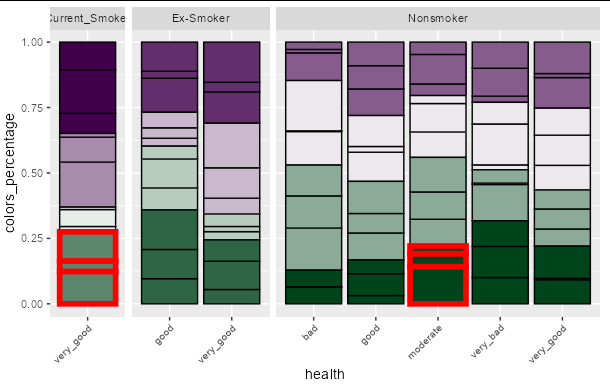
I would like to highlight just a few of the boxes that follow a certain condition in my geom_bar() plot by making their outlines thicker.
Labels for my conditions are:
colors = c('a','b','c','d','e','f','g','h','i','j','k','l')
health = c('very_good', 'good', 'moderate', 'bad', 'very_bad')
smoking_status = c('Current_Smoker','Ex-Smoker','Nonsmoke')
Code for my plot is:
smoking_health %>%
ggplot(aes(health, colors_percentage, fill = colors))
geom_bar(stat = 'identity', color = 'black')
facet_grid(~ smoking_status, scale = 'free', space = 'free_x')
scale_fill_manual(values = palette)
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust = 1, size = 8))
I want to highlight the conditions:
(smoking_status == 'Nonsmoke' & health == 'Moderate' & (colors %in% c('c', 'f'))) |
(smoking_status == 'Current_Smoker' & health == 'very_bad' & colors == 'e')
This is what I want for my final plot:
I can't seem to find a way to bolden the outline of these kind of specific conditions.
Does anyone know a way?
Thanks in advance!
CodePudding user response:
We don't have your data to reproduce your plot, but this approach should work for you. It relies on the fact that geom_bar can be given a size aesthetic to determine the width of its outline. You can create a new column in your data frame based on the logical rules you wish to use to highlight specific boxes, and feed this to the size parameter inside aes. Remember to add a scale_size_manual call to choose the width of the lines you want to use for normal and highlighted boxes.
smoking_health %>%
mutate(highlighted = (smoking_status == 'Nonsmoker' &
health == 'moderate' & colors %in% c("l", "a")) |
(smoking_status == 'Current_Smoker' & health == "very_good" &
colors == "j")) %>%
ggplot(aes(health, colors_percentage, fill = colors))
geom_bar(aes(size = highlighted),
stat = 'identity', color = 'black', position = "fill")
facet_grid(~ smoking_status, scale = 'free', space = 'free_x')
scale_fill_manual(values = palette)
scale_size_manual(values = c(0.5, 2))
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust = 1, size = 8))
theme(legend.position = "none")
Note that you can easily add a color scale using the same trick to make the boxes stand out even more:
smoking_health %>%
mutate(highlighted = (smoking_status == 'Nonsmoker' &
health == 'moderate' & colors %in% c("l", "a")) |
(smoking_status == 'Current_Smoker' & health == "very_good" &
colors == "j")) %>%
ggplot(aes(health, colors_percentage, fill = colors))
geom_bar(aes(size = highlighted, color = highlighted),
stat = 'identity', position = "fill")
facet_grid(~ smoking_status, scale = 'free', space = 'free_x')
scale_fill_manual(values = palette)
scale_size_manual(values = c(0.5, 2))
scale_color_manual(values = c("black", "red"))
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust = 1, size = 8))
theme(legend.position = "none")
Data used
smoking_health <- structure(list(colors_percentage = c(
0.424281163606793, 0.757145850220695,
0.272088683908805, 0.702225066488609, 0.783901083283126, 0.31812732364051,
0.25504200020805, 0.150501632830128, 0.949092260561883, 0.176580579485744,
0.597438448341563, 0.490371913416311, 0.310657090973109, 0.690649163676426,
0.815282948082313, 0.119425331009552, 0.112637027399614, 0.48738272418268,
0.628815256524831, 0.691388996317983, 0.516097098588943, 0.349926907802001,
0.894879485014826, 0.884916280163452, 0.683286903426051, 0.560128865065053,
0.87698360090144, 0.875387869309634, 0.62485910882242, 0.182680936297402,
0.827586045023054, 0.641206950880587, 0.920631993561983, 0.156408043345436,
0.696290905354545, 0.862210909835994, 0.59949598624371, 0.627007181989029,
0.671079852851108, 0.68094572937116, 0.52527018962428, 0.232511073350906,
0.671282195718959, 0.572535041952506, 0.00735216797329485, 0.217277518706396,
0.715195999480784, 0.998276757542044, 0.357245847582817, 0.645074530970305,
0.643338346853852, 0.0380584141239524, 0.0638308054767549, 0.674578275764361,
0.176417721668258, 0.0331670793239027, 0.21982069965452, 0.650513555621728,
0.303580678766593, 0.11516772210598, 0.0128057508263737, 0.114486665232107,
0.598777604289353, 0.759205947630107, 0.581150981364772, 0.649594946065918,
0.290496403817087, 0.772731678327546, 0.57460440415889, 0.474307884462178,
0.421877446817234, 0.638829972362146, 0.0785523615777493, 0.345375114353374,
0.187747624702752, 0.545154723571613, 0.309543700655922, 0.891319246962667,
0.72464850358665, 0.528383505064994, 0.0506025629583746, 0.286434431327507,
0.81406940240413, 0.920075967442244, 0.136473591905087, 0.990138751687482,
0.950963140232489, 0.32558137900196, 0.614674870390445, 0.881066406844184,
0.181831295602024, 0.8991091218777, 0.937552833463997, 0.277483612066135,
0.986416225554422, 0.626976334722713), colors = c("a", "b", "c",
"f", "i", "k", "l", "c", "d", "e", "f", "i", "l", "b", "c", "f",
"g", "h", "i", "l", "c", "e", "f", "i", "j", "k", "l", "c", "f",
"h", "i", "l", "a", "b", "c", "f", "i", "k", "l", "c", "d", "e",
"f", "i", "l", "b", "c", "f", "g", "h", "i", "l", "c", "e", "f",
"i", "j", "k", "l", "c", "f", "h", "i", "l", "a", "b", "c", "f",
"i", "k", "l", "c", "d", "e", "f", "i", "l", "b", "c", "f", "g",
"h", "i", "l", "c", "e", "f", "i", "j", "k", "l", "c", "f", "h",
"i", "l"), smoking_status = c("Current_Smoker", "Ex-Smoker",
"Nonsmoker", "Nonsmoker", "Nonsmoker", "Ex-Smoker", "Nonsmoker",
"Nonsmoker", "Current_Smoker", "Ex-Smoker", "Nonsmoker", "Nonsmoker",
"Nonsmoker", "Ex-Smoker", "Nonsmoker", "Nonsmoker", "Current_Smoker",
"Ex-Smoker", "Nonsmoker", "Nonsmoker", "Nonsmoker", "Ex-Smoker",
"Nonsmoker", "Nonsmoker", "Current_Smoker", "Ex-Smoker", "Nonsmoker",
"Nonsmoker", "Nonsmoker", "Ex-Smoker", "Nonsmoker", "Nonsmoker",
"Current_Smoker", "Ex-Smoker", "Nonsmoker", "Nonsmoker", "Nonsmoker",
"Ex-Smoker", "Nonsmoker", "Nonsmoker", "Current_Smoker", "Ex-Smoker",
"Nonsmoker", "Nonsmoker", "Nonsmoker", "Ex-Smoker", "Nonsmoker",
"Nonsmoker", "Current_Smoker", "Ex-Smoker", "Nonsmoker", "Nonsmoker",
"Nonsmoker", "Ex-Smoker", "Nonsmoker", "Nonsmoker", "Current_Smoker",
"Ex-Smoker", "Nonsmoker", "Nonsmoker", "Nonsmoker", "Ex-Smoker",
"Nonsmoker", "Nonsmoker", "Current_Smoker", "Ex-Smoker", "Nonsmoker",
"Nonsmoker", "Nonsmoker", "Ex-Smoker", "Nonsmoker", "Nonsmoker",
"Current_Smoker", "Ex-Smoker", "Nonsmoker", "Nonsmoker", "Nonsmoker",
"Ex-Smoker", "Nonsmoker", "Nonsmoker", "Current_Smoker", "Ex-Smoker",
"Nonsmoker", "Nonsmoker", "Nonsmoker", "Ex-Smoker", "Nonsmoker",
"Nonsmoker", "Current_Smoker", "Ex-Smoker", "Nonsmoker", "Nonsmoker",
"Nonsmoker", "Ex-Smoker", "Nonsmoker", "Nonsmoker"), health = c("very_good",
"good", "moderate", "very_good", "bad", "very_good", "good",
"very_bad", "very_good", "good", "moderate", "very_good", "bad",
"very_good", "good", "very_bad", "very_good", "good", "moderate",
"very_good", "bad", "very_good", "good", "very_bad", "very_good",
"good", "moderate", "very_good", "bad", "very_good", "good",
"very_bad", "very_good", "good", "moderate", "very_good", "bad",
"very_good", "good", "very_bad", "very_good", "good", "moderate",
"very_good", "bad", "very_good", "good", "very_bad", "very_good",
"good", "moderate", "very_good", "bad", "very_good", "good",
"very_bad", "very_good", "good", "moderate", "very_good", "bad",
"very_good", "good", "very_bad", "very_good", "good", "moderate",
"very_good", "bad", "very_good", "good", "very_bad", "very_good",
"good", "moderate", "very_good", "bad", "very_good", "good",
"very_bad", "very_good", "good", "moderate", "very_good", "bad",
"very_good", "good", "very_bad", "very_good", "good", "moderate",
"very_good", "bad", "very_good", "good", "very_bad")), row.names = c(1L,
2L, 3L, 6L, 9L, 11L, 12L, 15L, 16L, 17L, 18L, 21L, 24L, 26L,
27L, 30L, 31L, 32L, 33L, 36L, 39L, 41L, 42L, 45L, 46L, 47L, 48L,
51L, 54L, 56L, 57L, 60L, 61L, 62L, 63L, 66L, 69L, 71L, 72L, 75L,
76L, 77L, 78L, 81L, 84L, 86L, 87L, 90L, 91L, 92L, 93L, 96L, 99L,
101L, 102L, 105L, 106L, 107L, 108L, 111L, 114L, 116L, 117L, 120L,
121L, 122L, 123L, 126L, 129L, 131L, 132L, 135L, 136L, 137L, 138L,
141L, 144L, 146L, 147L, 150L, 151L, 152L, 153L, 156L, 159L, 161L,
162L, 165L, 166L, 167L, 168L, 171L, 174L, 176L, 177L, 180L), class = "data.frame")
palette <- c("#40004B", "#622E6B", "#855C8C", "#A88BAD", "#CAB9CD", "#EDE7EE",
"#E7EEEA", "#B9CCC0", "#8BAA97", "#5C876D", "#2E6544", "#00441B")