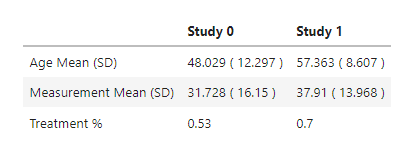
I have a dataset showing the distribution of variables between 2 studies.
library(knitr)
library(IRdisplay)
library(tidyr)
treat<- c(0,1,1,1,1,1) study<- c(0,1,0,0,0,0)
age<-c(38.07647, 35.30403, 42.19468, 28.72244, 38.84273, 28.74006)
measure<- c(36.36798, 51.21708, 37.71801, 38.84021, 26.70908, 36.39133)
df<- data.frame(treat,study,age,measure)
The goal is to recreate this table:
I have code below and was able to create the table but is there a way to refactor it to make it shorter/more readable? I'd like to add a function {} ideally but I'm not sure how to go about doing that for this code. This is what I wrote to recreate the table:
a<-df%>%
filter(study==0)%>%
summarise(study0=paste(round(mean(age),3), '(', round(sd(age),3),')', sep=""))
b<- df%>%
filter(study==1)%>%
summarise(study1=paste(round(mean(age),3), '(', round(sd(age),3),')', sep=""))
abbind<- bind_cols(a,b)
c<-df%>%
filter(study==0)%>%
summarise(study0=paste(round(mean(measure),3), '(', round(sd(measure),3),')', sep=""))
d<- df%>%
filter(study==1)%>%
summarise(study1=paste(round(mean(measure),3), '(', round(sd(measure),3),')', sep=""))
cdbind<- bind_cols(c,d)
e<- df%>%
filter(study==0)%>%
summarise(study0=paste(round((1-(sum(treat)/(nrow(df)-sum(treat)))), 2), sep=""))
f<- df%>%
filter(study==1)%>%
summarise(study1=paste(sum(treat)/sum(study), sep=""))
efbind<- bind_cols(e,f)
fullbind<- rbind(abbind, cdbind, efbind)
colnames(fullbind)<- c("Study 0", "Study 1")
rownames(fullbind)<-c('Age Mean (SD)', 'Measurement Mean (SD)', 'Treatment %')
Anyway to refactor this? Thank you guys for all your help!
CodePudding user response:
You can use this:
summ_table <- function(df) {
df %>%
mutate(study = paste("Study", study)) %>%
group_by(study) %>%
summarize(`Age mean (SD)` = paste(round(mean(age), 3), "(", round(sd(age), 3), ")"),
`Measurement mean (SD)` = paste(round(mean(measure), 3), "(", round(sd(measure), 3), ")"),
`Treatment %` = mean(treat)) %>%
gather("", value, -study) %>%
spread(study, value)
}
CodePudding user response:
This may be done with a group_by approach instead of filtering each value
library(dplyr)
library(tidyr)
library(stringr)
library(tibble)
df %>%
group_by(study) %>%
summarise(Treat = as.character(round(mean(treat))),
across(c(age, measure), ~ sprintf('%.2f (%.2f)',
mean(., na.rm = TRUE), sd(., na.rm = TRUE)),
.names = '{tools::toTitleCase(.col)} Mean (SD)')) %>%
pivot_longer(cols = -study) %>%
mutate(study = str_c('Study ', study)) %>%
pivot_wider(names_from = study, values_from = value) %>%
column_to_rownames('name')
-output
Study 0 Study 1
Treat 1 1
Age Mean (SD) 35.32 (6.21) 35.30 (NA)
Measure Mean (SD) 35.21 (4.86) 51.22 (NA)
-OP's fullbind output
> fullbind
Study 0 Study 1
Age Mean (SD) 35.315(6.207) 35.304(NA)
Measurement Mean (SD) 35.205(4.86) 51.217(NA)
Treatment % -1 1
data
df <- structure(list(treat = c(0, 1, 1, 1, 1, 1), study = c(0, 1, 0,
0, 0, 0), age = c(38.07647, 35.30403, 42.19468, 28.72244, 38.84273,
28.74006), measure = c(36.36798, 51.21708, 37.71801, 38.84021,
26.70908, 36.39133)), class = "data.frame", row.names = c(NA,
-6L))