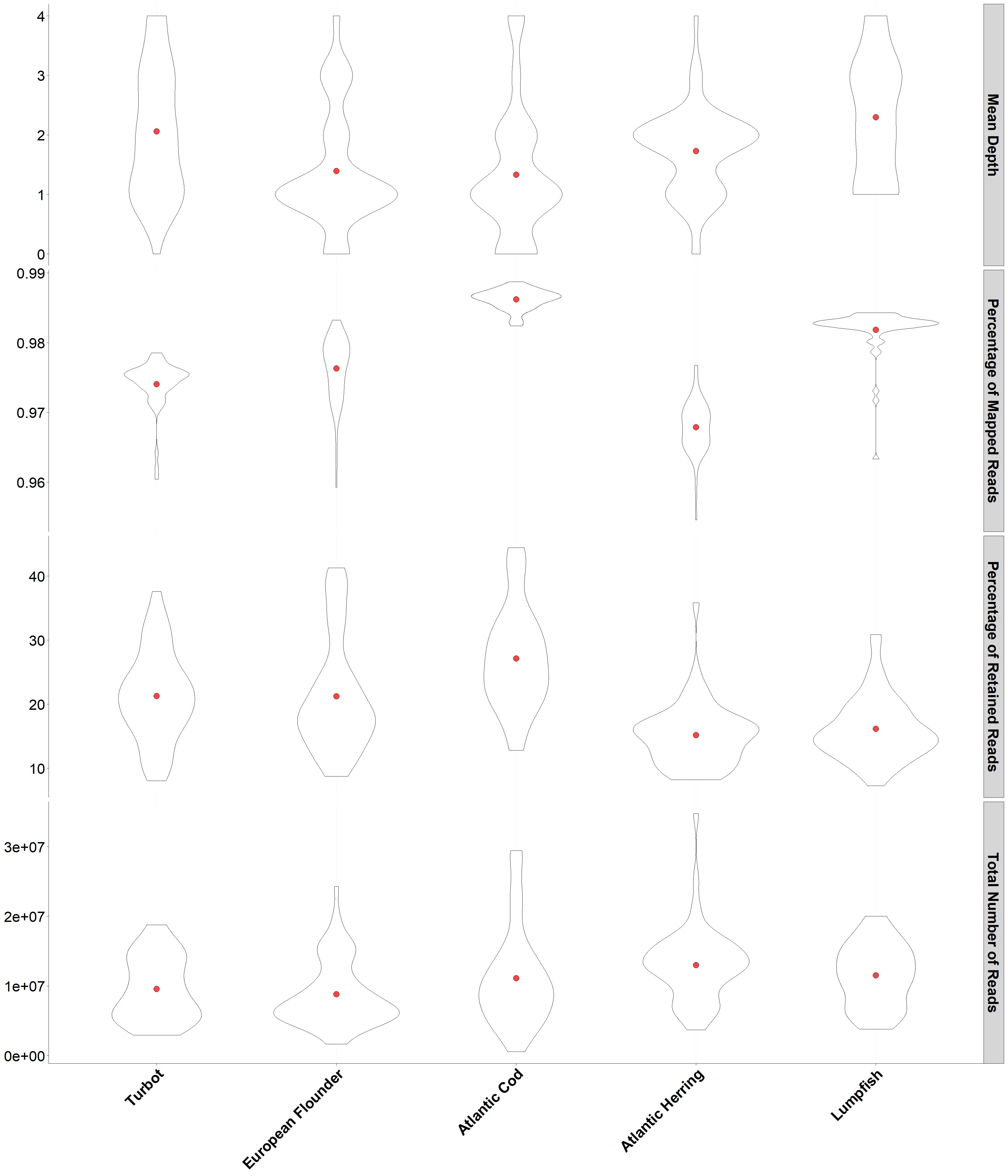
I have the ggplot2 code below that produces the plot attached. I would like to know if it would be possible to control the individual y-axis breaks/labels. For instance, I would like to have them for the Mean Depth as "1X", "2X" "3X" and "4X" & for Percentage of Retained Reads I would like to have "10%", "20%", "30%" and "40%". I guess I would need to use something like scale_y_continuous for each individual facet, but I have not been able to find a way of doing so.
Many thanks in advance, George.
ggplot()
geom_violin(data = fulldfUp, aes(x = Species, y = Value),
fill = "#ffffff", colour = "#000000", show.legend = FALSE, alpha = .9, size = .3, width = .7)
stat_summary(data = fulldfUp, aes(x = Species, y = Value),
fun = mean, geom = "point", shape = 21, size = 3.5, alpha = .9, colour = "#000000", fill = "#fc3232")
facet_grid(Estimate ~. , scales = "free", labeller = labeller(Estimate = ylabels))
scale_fill_manual(values = c("#fbb4ae", "#b3cde3", "#ccebc5", "#decbe4", "#fed9a6"))
scale_colour_manual(values = c("#fbb4ae", "#b3cde3", "#ccebc5", "#decbe4", "#fed9a6"))
theme(panel.background = element_rect(fill = "#ffffff"),
panel.grid.major.x = element_line(color = "#ededed", linetype = "dashed", size = .00005),
panel.grid.major.y = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
axis.line = element_line(colour = "#000000", size = .3),
axis.title = element_blank(),
axis.text.x = element_text(colour = "#000000", size = 20, face = "bold", angle = 45, vjust = 1, hjust = 1),
axis.text.y = element_text(color = "#000000", size = 20),
axis.ticks.x = element_line(color = "#000000", size = .3),
axis.ticks.y = element_line(color = "#000000", size = .3),
strip.background.y = element_rect(colour = "#000000", fill = "#d6d6d6", size = 0.3),
strip.text = element_text(colour = "#000000", size = 20, face = "bold"),
legend.position = "top",
legend.margin = margin(t = 0, b = 0, r = 0, l = 0),
legend.box.margin = margin(t = 10, b = 20, r = 0, l = 0),
legend.key = element_rect(fill = NA),
legend.background = element_blank())
CodePudding user response:
I needed to create my own example data because it was not provided.
You can normalize the data per group to plot the percentages:
library(tidyverse)
set.seed(1337)
# simulate some aboundance data
data <-
list(
Species_a = rpois(100, 0.2),
Species_b = rpois(100, 0.1),
Species_c = rpois(100, 0.3),
Species_d = rpois(100, 0.8),
Species_e = rpois(100, 0.7)
) %>%
enframe() %>%
unnest(value) %>%
mutate(group = ifelse(name %in% c("Species_d", "Species_e"), "B", "A"))
data
#> # A tibble: 500 x 3
#> name value group
#> <chr> <int> <chr>
#> 1 Species_a 0 A
#> 2 Species_a 0 A
#> 3 Species_a 0 A
#> 4 Species_a 0 A
#> 5 Species_a 0 A
#> 6 Species_a 0 A
#> 7 Species_a 1 A
#> 8 Species_a 0 A
#> 9 Species_a 0 A
#> 10 Species_a 0 A
#> # … with 490 more rows
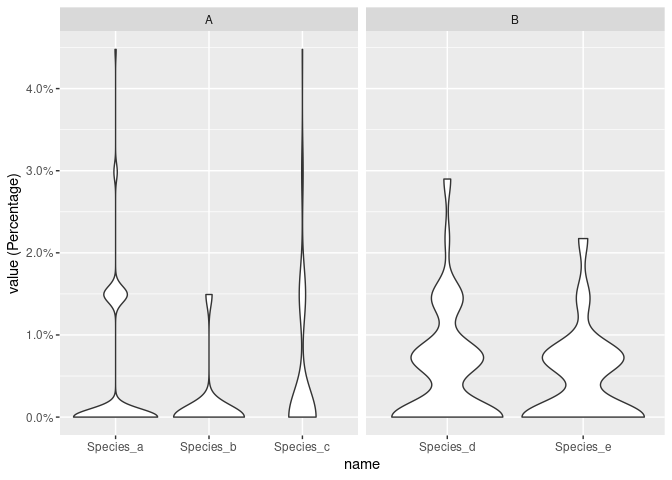
data %>%
group_by(group) %>%
mutate(value = value / sum(value) * 100) %>%
ggplot(aes(name, value))
geom_violin()
facet_wrap(~group, scales = "free_x")
scale_y_continuous(labels = scales::percent_format(scale = 1))
labs(y = "value (Percentage)")