Based on the output dataframe from 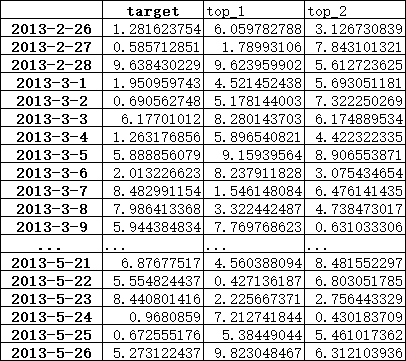
Pseudocode:
df2 = df.filter(regex='_mape$').groupby(pd.Grouper(freq='M')).dropna(axis=1, how='all')
df2.columns = ['top_1', 'top_2', ..., 'top_k']
df.join(df2)
As @Quang Hoang commented in the last post, we may could use justify_nd to achieve that, but I don't know how. Thanks for your help at advance.
CodePudding user response:
Actually, what you need is, for each group (by year / month):
- compute errors locally for the current group,
- find k "wanted" columns (calling argsort) and take indicated columns from models,
- take the indicated columns from the current group and rename them to top_…,
- return what you generated so far.
To do it, define a "group processing" function:
def grpProc(grp):
err = mape(grp[models], grp[['target']])
sort_args = np.argsort(err) < k
cols = models[sort_args]
out_cols = [f'top_{i 1}' for i in range(k)]
rv = grp.loc[:, cols]
rv.columns=out_cols
return rv
Then, to generate top_… columns alone, apply this function to each group:
wrk = df.groupby(pd.Grouper(freq='M')).apply(grpProc)
And finally generate the expected result joining target column with wrk:
result = df[['target']].join(wrk)
Edit
For the first group (2013-02-28) err contains:
A_values 48.759348
B_values 77.023855
C_values 325.376455
D_values 74.422508
E_values 60.602101
Note that 2 lowest error values are 48.759348 and 60.602101, so from this group you should probably take A_values (this is OK) and E_values (instead of D_values).
So maybe grpProc function instead of:
sort_args = np.argsort(err) < k
cols = models[sort_args]
should contain:
cols = err.nsmallest(k).index
