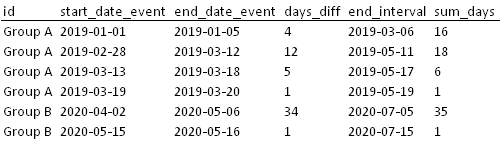
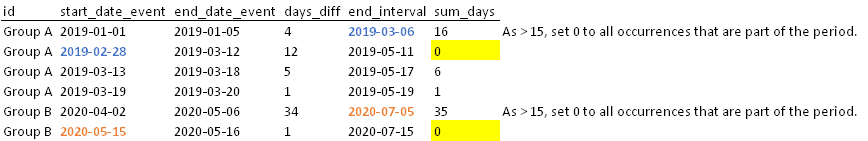
I have the following dataset.
Based on the "start_date_event" of each row, I have already summed up all the days that occurred within a 60 day range (variable sum_days) from the respective event start date.
However there is a condition that only sums greater than 15 days, for example, must be considered. Therefore, for occurrences longer than 15 days I would like to assign "0" to all rows that are part of that respective period.
Expected output:
Example of expected result: Row 2 has become 0 as it is contained within the range of the previous row whose sum is greater than fifteen days. The event recorded in row 2 starts on 2019-02-28, which belongs to the period 2019-01-01 (start of the event) to 2019-03-06 (end of the 60-day interval, 01-01-2019 60) of the first row whose sum is greater than 15.
Does anyone have any suggestions?
Reproducible example:
library(data.table)
library(dplyr)
# Input data
data <- data.table(id = c("Group A", "Group A", "Group A", "Group A",
"Group B", "Group B"),
start_date_event = c("2019-01-01",
"2019-02-28",
"2019-03-13",
"2019-03-19",
"2020-04-02",
"2020-05-15"),
end_date_event = c("2019-01-05",
"2019-03-12",
"2019-03-18",
"2019-03-20",
"2020-05-06",
"2020-05-16"))
# Convert to date
data <- data %>%
dplyr::mutate(start_date_event = as.Date(start_date_event)) %>%
dplyr::mutate(end_date_event = as.Date(end_date_event)) %>%
dplyr::mutate(days_diff = as.integer(end_date_event - start_date_event)) %>%
dplyr::mutate(end_interval = end_date_event 60) %>%
data.table::setDT()
# Calculating cumulative sum within 60 days
data[.(c = id, tmin = start_date_event,
tmax = start_date_event 60),
on = .(id == c, start_date_event <= tmax,
start_date_event >= tmin),
sum_days := sum(days_diff), by = .EACHI]
CodePudding user response:
This should work:
library(sqldf)
library(dplyr)
library(data.table)
# Creating a new 'row column'
data$row_n <- 1:nrow(data)
# Identifying which lines overlap and then filtering data
data <- sqldf("select a.*,
coalesce(group_concat(b.rowid), '') as overlaps
from data a
left join data b on a.id = b.id and
not a.rowid = b.rowid and
((a.start_date_event between
b.start_date_event and b.end_interval) or
(b.start_date_event between a.start_date_event
and a.end_interval))
group by a.rowid
order by a.rowid") %>%
group_by(id) %>%
mutate(row_n = as.character(row_n),
previous_row = dplyr::lag(row_n, n = 1, default = NA),
previous_value = dplyr::lag(sum_days, n = 1, default = NA),
sum2 = case_when(mapply(grepl,previous_row, overlaps) == TRUE &
previous_value > 15 ~ as.integer(0),
TRUE ~ sum_days),
previous_value = dplyr::lag(sum2, n = 1, default = NA),
sum2 = case_when(mapply(grepl,previous_row, overlaps) == TRUE &
previous_value > 15 ~ as.integer(0),
TRUE ~ sum_days)) %>%
dplyr::select(-c(previous_value, previous_row, row_n))