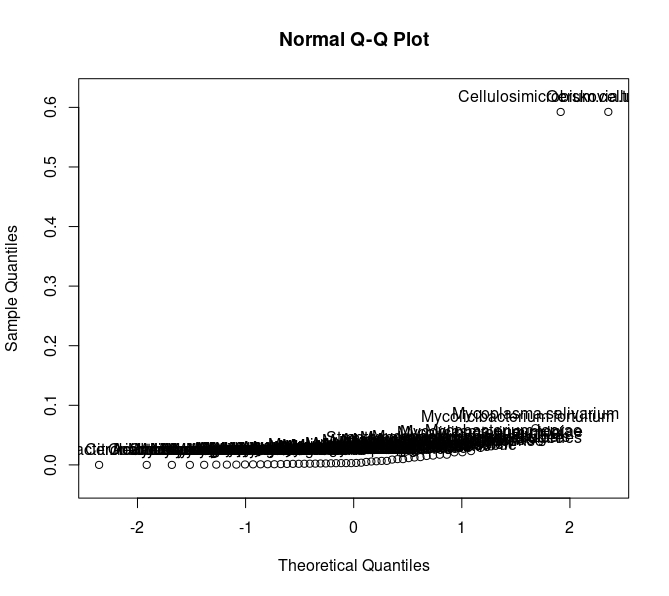
I'm trying to generate a QQ plot where I would like to label the respective points.
This is my data subset
dput(head(C_A,10))
structure(list(CONTROL_ASYMPTOMATIC = structure(c(3L, 4L, 1L,
53L, 5L, 11L, 35L, 31L, 20L, 45L), .Label = c("Acidocella.facilis",
"Cellulosimicrobium.cellulans", "Citrobacter.freundii", "Citrobacter.koseri",
"Gallibacterium.anatis", "Mycobacterium.asiaticum", "Mycobacterium.avium",
"Mycobacterium.cookii", "Mycobacterium.gastri", "Mycobacterium.gordonae",
"Mycobacterium.hyorhinis", "Mycobacterium.intracellulare", "Mycobacterium.kansasii",
"Mycobacterium.leprae", "Mycobacterium.malmoense", "Mycobacterium.marinum",
"Mycobacterium.scrofulaceum", "Mycobacterium.simiae", "Mycobacterium.sp.",
"Mycobacterium.szulgai", "Mycobacterium.tuberculosis", "Mycobacterium.ulcerans",
"Mycobacterium.xenopi", "Mycobacteroides.chelonae", "Mycolicibacillus.trivialis",
"Mycolicibacterium.aichiense", "Mycolicibacterium.aurum", "Mycolicibacterium.chitae",
"Mycolicibacterium.chubuense", "Mycolicibacterium.diernhoferi",
"Mycolicibacterium.fallax", "Mycolicibacterium.farcinogenes",
"Mycolicibacterium.flavescens", "Mycolicibacterium.fortuitum",
"Mycolicibacterium.gadium", "Mycolicibacterium.gilvum", "Mycolicibacterium.komossense",
"Mycolicibacterium.neoaurum", "Mycolicibacterium.obuense", "Mycolicibacterium.phlei",
"Mycolicibacterium.senegalense", "Mycolicibacterium.smegmatis",
"Mycolicibacterium.sphagni", "Mycolicibacterium.thermoresistibile",
"Mycolicibacterium.vaccae", "Mycoplasma.agalactiae", "Mycoplasma.capricolum",
"Mycoplasma.hyorhinis", "Mycoplasma.neurolyticum", "Mycoplasma.pneumoniae",
"Mycoplasma.salivarium", "Oerskovia.turbata", "Orientia.tsutsugamushi",
"Streptosporangium.amethystogenes"), class = "factor"), p_value = c(7.75e-07,
7.83e-07, 1.72e-06, 2.89e-05, 0.000133449, 0.000419124, 0.000422792,
0.000452307, 0.000726928, 0.000914429)), row.names = c(NA, 10L
), class = "data.frame")
To generate the QQ Plot
My code
loc <- qqnorm(C_A$p_value, ylim = extendrange(C_A$p_value, f = 0.05))
text(loc[[1]], loc[[2]], C_A$CONTROL_ASYMPTOMATIC, pos = 3)
Is there any way to improve the labeling such as only those points which have the highest significance.
Any suggestion or help would be really appreciated
CodePudding user response:
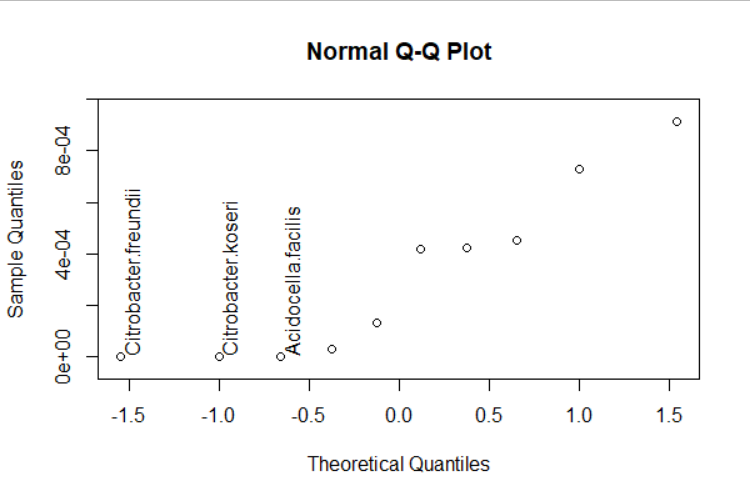
Subset significant ones when adding the text:
#plot dots
loc <- qqnorm(C_A$p_value, ylim = extendrange(C_A$p_value, f = 0.05))
#get the index of significant points
#note: I am using 0.00001 as significant,
# amend this number as per your needs.
ix <- which(C_A$p_value < 0.00001)
#subset and add text
text(loc[[1]][ ix ], loc[[2]][ ix ],
C_A$CONTROL_ASYMPTOMATIC[ ix ], pos = 4, srt = 90)