I'm trying to show the growth of COVID cases in New York state
This code gets the plot I want but without the animation or aspect of time.
Full error:
Error in insert_points(polygon$x, polygon$y, splits, n):
Not compatible with requested type: [type=NULL; target=double].
county_map = map_data("county", region = "New York")
county_map$region = county_map$subregion
covidCounties = read.csv("https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-counties.csv", header = T)
covidCounties = covidCounties %>%
mutate(date = as.Date(date)) %>%
filter(state == "New York") %>%
arrange(date)%>%
group_by(county) %>%
mutate(county = tolower(county)) %>%
mutate(newCases = diff(c(0, cases))) %>%
mutate(newDeaths = diff(c(0, deaths))) %>%
ungroup() %>%
select(date, state, county, cases, newCases, deaths)
covidCountyMap = covidCounties %>%
ggplot(aes(
map_id = county,
fill = newCases,
group = county
))
geom_map(
map = county_map,
color = "black"
)
expand_limits(x = county_map$long, y = county_map$lat)
scale_fill_gradientn(colors = c("green", "yellow", "red"), breaks = c(0, 100, 500))
labs(
title = "New cases over time in New York State",
subtitle = "{frame_time}"
)
covidCountyMap
covidCountyMap
transition_time(date)
CodePudding user response:
You need to tell {gganimate} what polygons to transition to one another. It won't be able to guess that for you. In other words, you need to add a group identifier to each transition state (meaning each county by date).
I filtered to only one state because the reprex on the entire data kept crashing. I have transformed to a log scale for your counts, in order to represent the data range better. (there are a few negative values, therefore the warning)
library(tidyverse)
library(gganimate)
county_map = map_data("county", region = "New York")
county_map$region = county_map$subregion
## I'd advise to create a separate data frame for your raw data, and not overwrite it
covidCounties_raw = read.csv("https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-counties.csv", header = T)
covidCounties <- covidCounties_raw %>%
mutate(date = as.Date(date)) %>%
filter(state == "New York") %>%
arrange(date) %>%
group_by(county) %>%
mutate(county = tolower(county)) %>%
mutate(newCases = diff(c(0, cases))) %>%
mutate(newDeaths = diff(c(0, deaths))) %>%
ungroup() %>%
select(date, state, county, cases, newCases, deaths) %>%
## this is the main trick
group_by(date, county) %>%
mutate(id = cur_group_id()) %>%
ungroup() %>%
## I'm filtering for only one county because the reprex took too long with the entire data
filter(county == "nassau")
covidCountyMap <- covidCounties %>%
ggplot(aes(
map_id = county,
fill = newCases,
## use the group identifier for your grouping
group = id
))
geom_map(
map = county_map,
color = "black"
)
expand_limits(x = county_map$long, y = county_map$lat)
scale_fill_gradientn(colors = c("green", "yellow", "red"),
## log transformed scale
trans = "log")
labs(
title = "New cases over time in New York State",
subtitle = "{frame_time}"
)
anim <- covidCountyMap
transition_time(date)
## have slightly reduced the frame rate to make it slightly faster
animate(anim, fps = 5, nframes = 50)
#> Warning: Transformation introduced infinite values in discrete y-axis
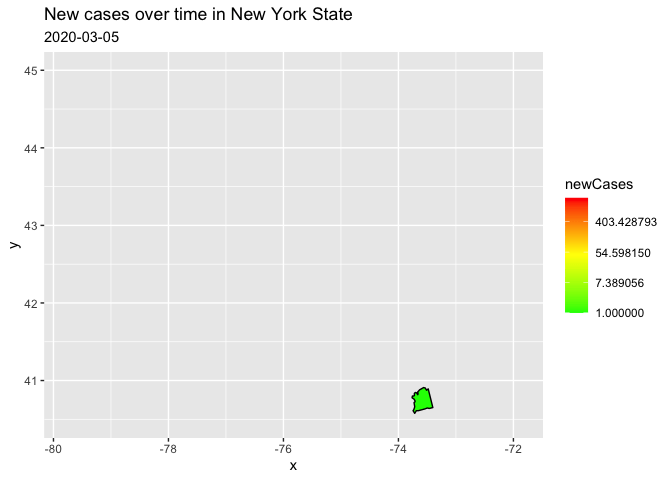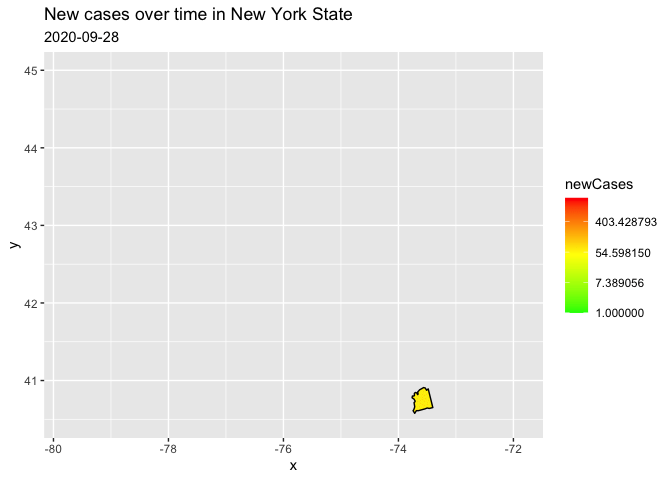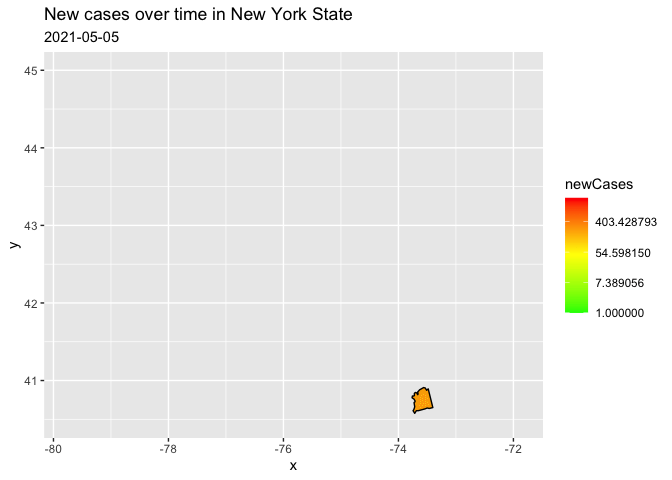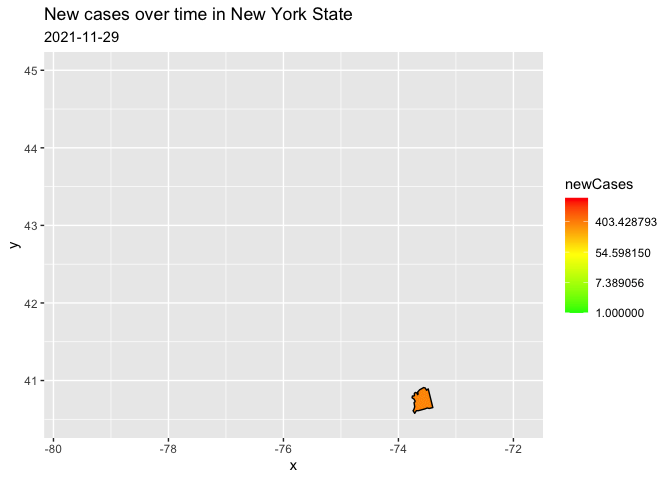
Created on 2021-11-30 by the reprex package (v2.0.1)
