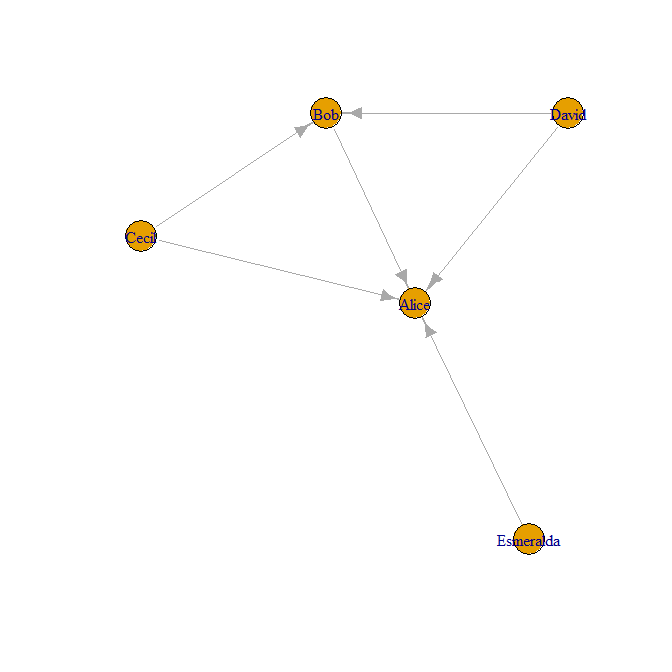
The following is an example taken from the igraph package:
library(igraph)
actors <- data.frame(name=c("Alice", "Bob", "Cecil", "David", "Esmeralda"))
relations <- data.frame(from=c("Bob", "Cecil", "Cecil", "David","David", "Esmeralda"),
to=c("Alice", "Bob", "Alice", "Alice", "Bob", "Alice"),
friendship=c(4,5,5,2,1,1))
g <- graph_from_data_frame(relations, directed=TRUE, vertices=actors)
The plot of the graph is the following:
However, I would like to get a different plot. The aim is to improve the quality and aesthetics of the graphic, for example by using ggplot2 (or other solutions which I don't know).
The plot should not contain links between nodes: the nodes should be closer to each other, the greater the weight of the link that exists between them. The nodes should have their own label (as depicted in the plot above) and their size should be proportional to the value of the variable p of the dataframe df.
> dput(df)
structure(list(nodes = c("Alice", "Bob", "Cecil", "David", "Esmeralda"
), p = c(4, 3, 2, 2, 1)), class = "data.frame", row.names = c(NA,
-5L))
> df
nodes p
1 Alice 4
2 Bob 3
3 Cecil 2
4 David 2
5 Esmeralda 1
CodePudding user response:
Update
Maybe you can use visNetwork package
library(visNetwork)
nodes <- transform(
setNames(df, c("label", "value")),
id = seq_along(label)
)
edges <- transform(
relations,
from = match(from, nodes$label),
to = match(to, nodes$label),
length = prod(friendship) / friendship
)
visNetwork(nodes, edges) %>%
visEdges(hidden = TRUE)
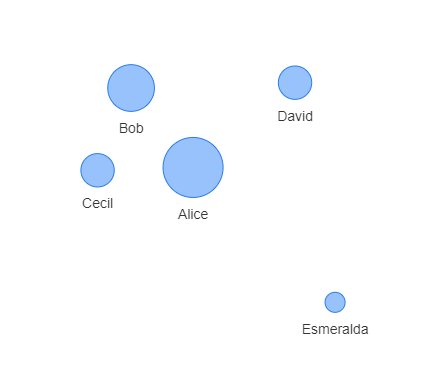
which gives
Previous Answer
Maybe you can try the code below
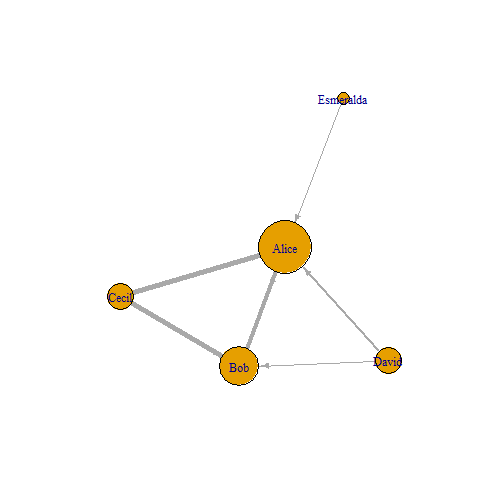
g %>%
set_edge_attr(name = "width", value = E(.)$friendship) %>%
set_vertex_attr(name = "size", value = 10 * with(df, p[match(nodes, names(V(.)))])) %>%
plot()
CodePudding user response:
One simple solution is to treat the edge value friendship as edge weight. Internally, these weights are in turn fed to into the standard layout algorithm of igraph (?layout_nicely; ?layout_with_fr see weights argument):
E(g)$weight <- E(g)$friendship^2 # squaring increases the observed effect
plot(g)
or as one-liner:
plot(g, layout = layout_with_fr(g, weights = E(g)$friendship^2)
To omit links you could simply set the color of edges to white:
plot(g,
layout = layout_with_fr(g, weights = E(g)$friendship^2,
edge.color = 'white'))
Or in a more elegant solution calculate the layout beforehand and delete the links altogether.
l <- layout_with_fr(g, weights = E(g)$friendship^2)
g <- delete.edges(g, which(E(g)$friendship > 0))
plot(g, layout = l)