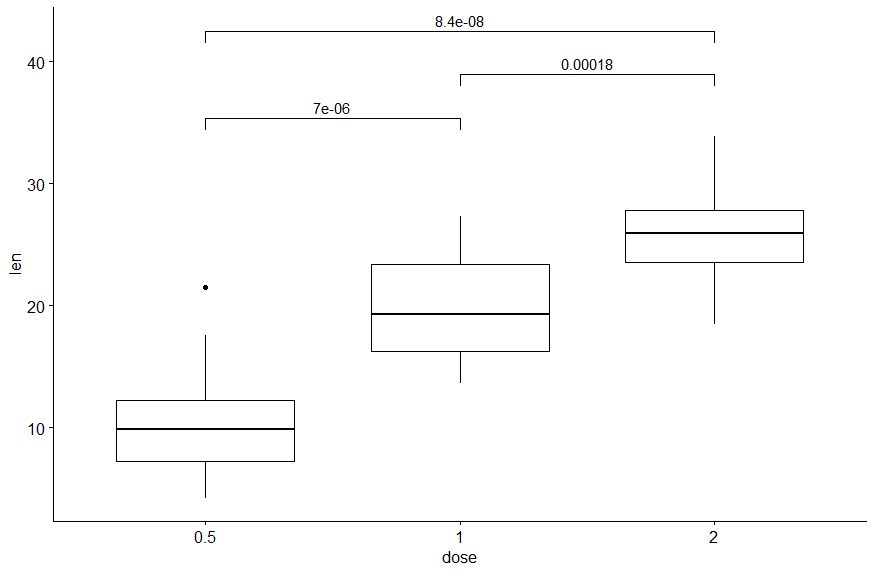I try to make boxplot with displaying p-value
my_comparisons <- list( c("0.5", "1"), c("1", "2"), c("0.5", "2") )
ggboxplot(ToothGrowth, x = "dose", y = "len")
stat_compare_means(comparisons = my_comparisons, method = "wilcox.test")
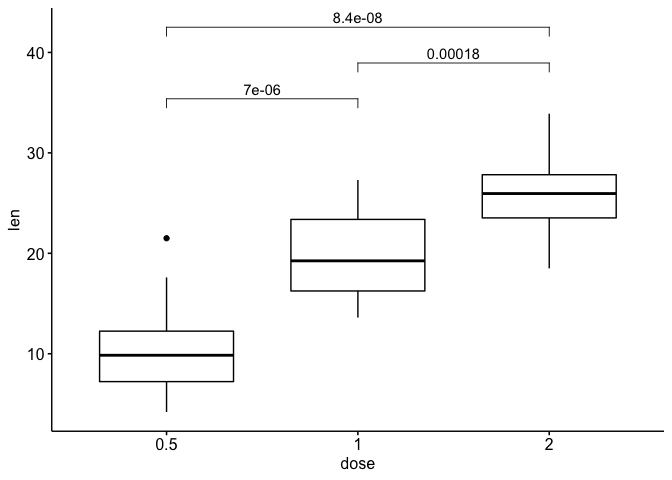
the result is
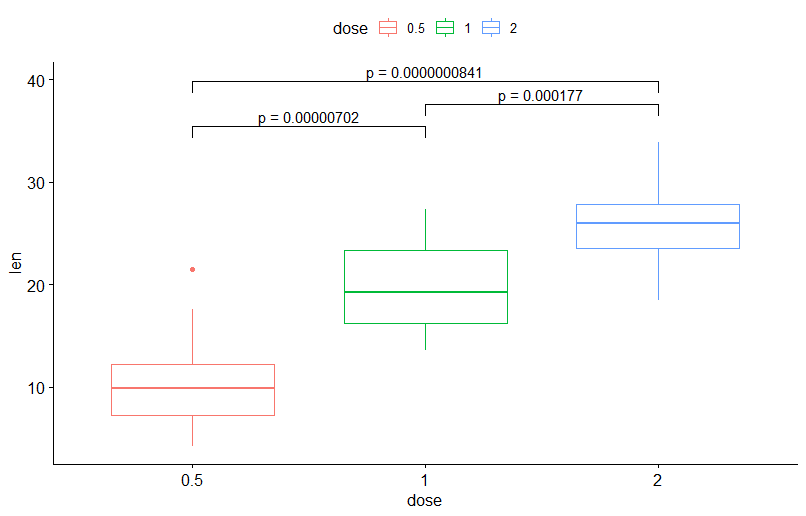
But how can I add additional text to calculated p-values? I whant to add "p = " as shown below
How can I make it?
upd. Variants below not working
my_comparisons <- list( c("0.5", "1"), c("1", "2"), c("0.5", "2") )
ggboxplot(ToothGrowth, x = "dose", y = "len")
stat_compare_means(comparisons = my_comparisons, method = "wilcox.test", aes(label=paste("p=",scientific(as.numeric(..p.format..)))))
ggboxplot(ToothGrowth, x = "dose", y = "len")
stat_compare_means(comparisons = my_comparisons, method = "wilcox.test", aes(label = paste("p =", ..p.format..)))
ggboxplot(ToothGrowth, x = "dose", y = "len")
stat_compare_means(comparisons = my_comparisons, method = "wilcox.test", aes(label = paste0("p =", ..p.format..)))
CodePudding user response:
I believe this could be potentially a bug in ggpubr. The documentation provides an example how to add custom text to your p-value. This starts to fail when passing several comparisons to the labelling function. I think the bug might be that ggplot doesn't know which expression to parse to a label (see r-lang warning below). There are also several warnings regarding the calculation of the p-value that might be relevant.
That said, I feel that most questions regarding ggpubr are concerning annotations with multiple p-values, and I do feel that you might want to reconsider (a) your statistics and (b) your visualisation.
library(ggpubr)
#> Loading required package: ggplot2
library(tidyverse)
## This works as expected. Only one test
ggboxplot(ToothGrowth, x = "supp", y = "len")
stat_compare_means(aes(label = paste0("p = ", ..p.format..)))
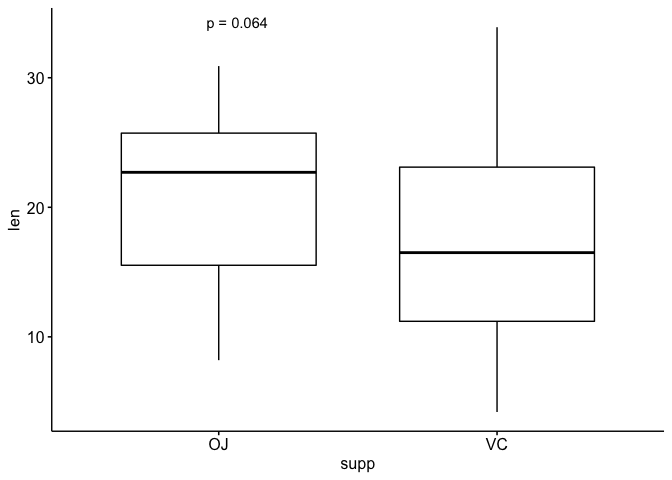
See plenty of warnings - starts to fail.
my_comparisons <- list( c("0.5", "1"), c("1", "2"), c("0.5", "2") )
ggboxplot(ToothGrowth, x = "dose", y = "len")
stat_compare_means(comparisons = my_comparisons,
aes(label = paste0("p = ", ..p.format..)))
#> Warning: Using `as.character()` on a quosure is deprecated as of rlang 0.3.0.
#> Please use `as_label()` or `as_name()` instead.
#> This warning is displayed once per session.
#> Warning in wilcox.test.default(c(4.2, 11.5, 7.3, 5.8, 6.4, 10, 11.2, 11.2, :
#> cannot compute exact p-value with ties
#> Warning in wilcox.test.default(c(4.2, 11.5, 7.3, 5.8, 6.4, 10, 11.2, 11.2, :
#> cannot compute exact p-value with ties
#> Warning in wilcox.test.default(c(16.5, 16.5, 15.2, 17.3, 22.5, 17.3, 13.6, :
#> cannot compute exact p-value with ties
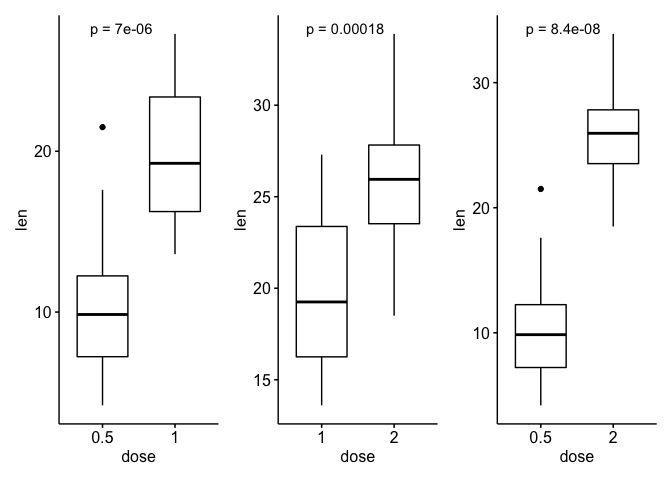
By creating a plot for each compared group separately, this starts to work again.
ls_tg <- ToothGrowth %>%
split(., .$dose)
lapply(my_comparisons, function(x) bind_rows(ls_tg[x])) %>%
map(~ggboxplot(., x = "dose", y = "len")
stat_compare_means(aes(label = paste0("p = ", ..p.format..)))) %>%
patchwork::wrap_plots()
To get more control over the format and rounding you could use this or alter to any format you wish.
mutate(myformatted.p = paste0("p = ", ifelse(p < 1e-5, format(p, scientific = T), signif(p, digits = 2))))