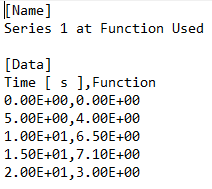
I am reading .csv files into R that were produced by software that adds extra labels to the data it exports, without placing commas following these extra labels. A simplified version of the text file can be seen in the following image.
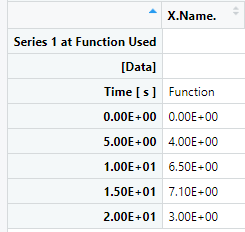
When I import using the read.csv() function and view the data I get the following:
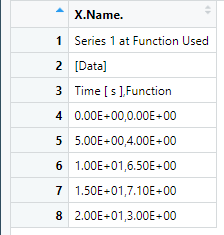
I then tried read.csv2, and wrote a new file removing the first two rows:
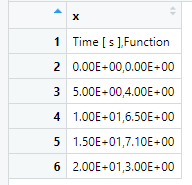
Modified to:
This is displayed in a text file as:
This is becoming a laborious task, and I am wondering whether there is a more efficient way of tidying up the files (as I have a lot of them).
Basically what I am trying to do is create a .csv file that when read into R has two columns with the first headed "Time [s]" and the second column headed "Function" The header "x" is not followed by a comma when exported as a new .csv file and is preventing me from reading my file into R in the form that I described.
The data can be copied and pasted (I think), using:
structure(list(X.Name. = c("", "", "Function", "0.00E 00", "4.00E 00", "6.50E 00", "7.10E 00", "3.00E 00")), class = "data.frame", row.names = c("Series 1 at Function Used", "[Data]", "Time [ s ]", "0.00E 00", "5.00E 00", "1.00E 01", "1.50E 01", "2.00E 01"))
CodePudding user response:
I think you're facing a common misconception of read_csv() that most people eventually come across when they begin programming in R.
There are a large number of arguments attached to the read family functions and they really need to be explored to understand the extent of {readr}.
In this specific problem, you have a very clear, 4 row jump before the headers of your data.
You can use the skip argument in the read_csv() function to skip 4 rows. Alternatively, let's say you wanted to change the name of your columns. You could also do this using different arguments within read_csv().
For further advice and functionality feel free to do ?read_csv() to look at the help file and has all the information around the different arguments for that function.
Relative to the amount of files that you have to "read in", I would recommend building your own function to automate the process.
Something like:
read_files <- function(my_file_paths) {
read_csv(my_file_paths, skip = 4....)
whatever else you want to do.....
}