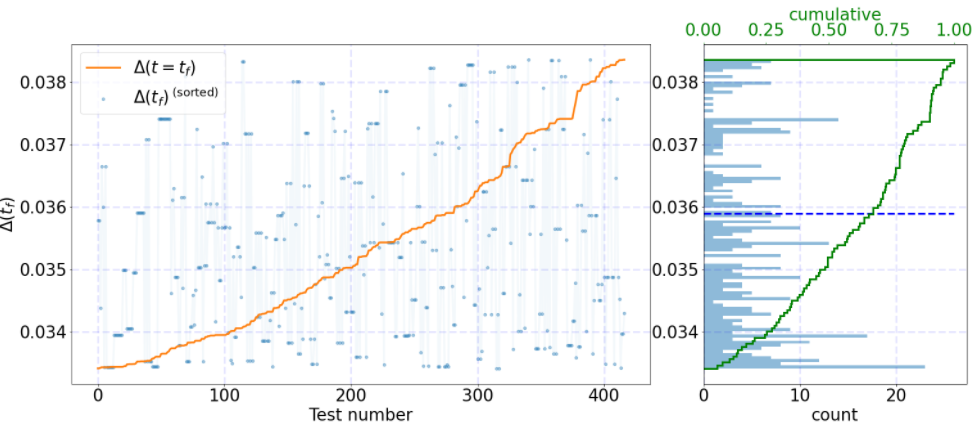
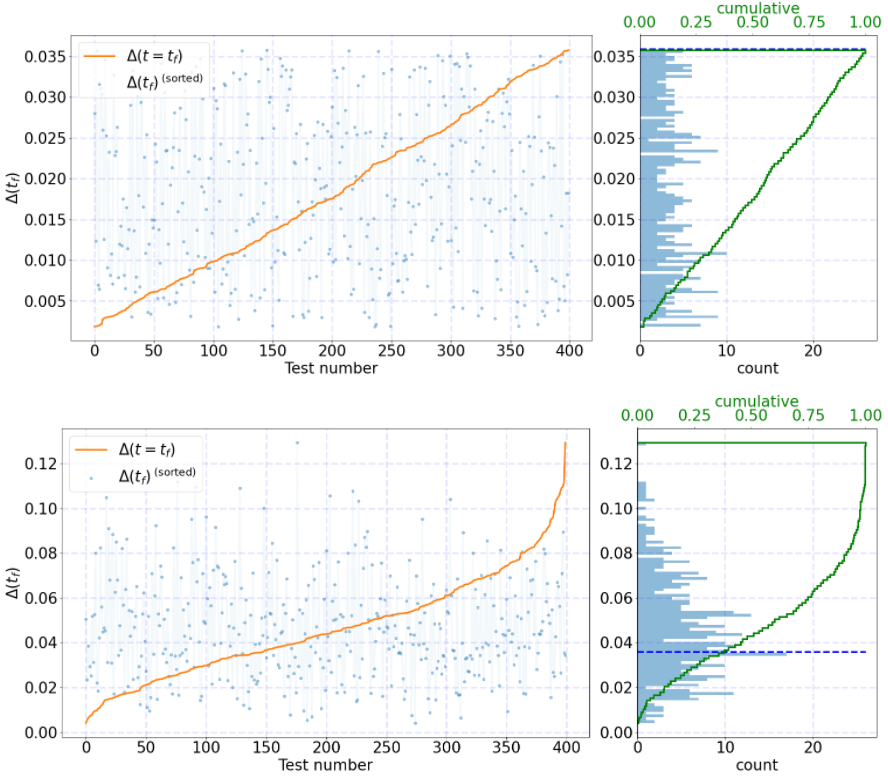
I'm trying to fix the format for this plot:
that I'm generating with "Df" data, but I'm having this cumbersome issue in which both plots are too close. The script I'm using to plot this is:
#[Df,mdD,teoD,e1f,teoe1,e2f,teoe2] = [[row[i] for row in np.load('master_results.npy',allow_pickle=True)[1][2:]] for i in [0,1,2,3,4,5,6]] #reading Datafile
import matplotlib.gridspec as gridspec
def test_histograms(values):
fig = plt.figure(figsize=(25,15))
gs = gridspec.GridSpec(3, 3)
ax_main = plt.subplot(gs[1:3, :2])
ax_yDist = plt.subplot(gs[1:3, 2],sharey=ax_main)
ax_main.plot(np.arange(len(values)),values, alpha=0.05)
ax_main.scatter(np.arange(len(values)),values, s=20, marker='o',alpha=0.4)
values.sort()
ax_main.plot(values, linewidth=3)
ax_main.set(xlabel="Test number", ylabel=r"$\Delta(t_f)$")
ax_main.grid(True, color='blue', linewidth=3, linestyle='--', alpha=0.1)
ax_main.legend(['_nolegend_', r'$\Delta(t=t_f)$', '$\Delta(t_f)^{\;\mathrm{(sorted)}}$'])
ax_yDist.hist(values,bins=100,orientation='horizontal',align='mid', alpha=0.5)
ax_yDist.hlines(0.0359,0,26,linestyle='--',color='blue')
ax_yDist.grid(True, color='blue', linewidth=3, linestyle='--', alpha=0.1)
ax_yDist.set(xlabel='count')
ax_yCumDist = ax_yDist.twiny()
ax_yCumDist.hist(values,bins=100,cumulative=True,histtype='step',density=True,color='green',align='mid',orientation='horizontal',linewidth=3)
ax_yCumDist.tick_params('x', colors='green')
ax_yCumDist.set_xlabel('cumulative',color='green')
plt.show()
test_histograms(Df)
Df=np.random.uniform(0.0359, 0.0018, 400)
test_histograms(Df)
Df=np.random.rayleigh(0.0359, 400)
test_histograms(Df)
Df=np.random.normal(0.0359, 0.0018, 400)
test_histograms(Df)
You can use different distributions to allow reproducibility. Specifically, for 3 decimal values, the 2nd (right) yaxis is too close from the left-hand. I would expect to see something like
the 2nd plot but for any range possible. How could I fix this? Maybe adding a thin "space" between left and right plots?
CodePudding user response:
Use the tight_layout() function to adjust the padding between and around subplots:
plt.tight_layout()
Code:
import numpy as np
from matplotlib import pyplot as plt
from matplotlib import gridspec as gridspec
def test_histograms(values):
fig = plt.figure(figsize=(25,15))
gs = gridspec.GridSpec(3, 3)
ax_main = plt.subplot(gs[1:3, :2])
ax_yDist = plt.subplot(gs[1:3, 2],sharey=ax_main)
_ = ax_main.plot(np.arange(len(values)),values, alpha=0.05)
ax_main.scatter(np.arange(len(values)),values, s=20, marker='o',alpha=0.4)
values.sort()
_ = ax_main.plot(values, linewidth=3)
ax_main.set(xlabel="Test number", ylabel=r"$\Delta(t_f)$")
ax_main.grid(True, color='blue', linewidth=3, linestyle='--', alpha=0.1)
ax_main.legend(['_nolegend_', r'$\Delta(t=t_f)$', '$\Delta(t_f)^{\;\mathrm{(sorted)}}$'])
ax_yDist.hist(values,bins=100,orientation='horizontal',align='mid', alpha=0.5)
ax_yDist.hlines(0.0359,0,26,linestyle='--',color='blue')
ax_yDist.grid(True, color='blue', linewidth=3, linestyle='--', alpha=0.1)
ax_yDist.set(xlabel='count')
ax_yCumDist = ax_yDist.twiny()
ax_yCumDist.hist(values,bins=100,cumulative=True,histtype='step',density=True,color='green',align='mid',orientation='horizontal',linewidth=3)
ax_yCumDist.tick_params('x', colors='green')
ax_yCumDist.set_xlabel('cumulative',color='green')
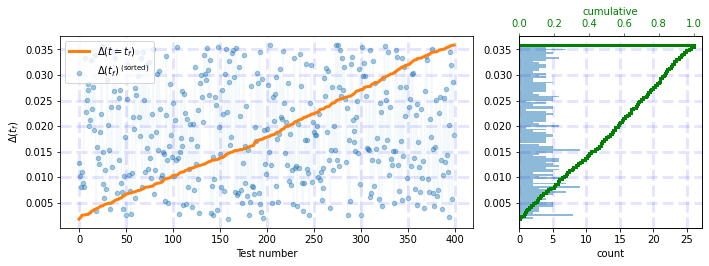
plt.tight_layout()
plt.show()
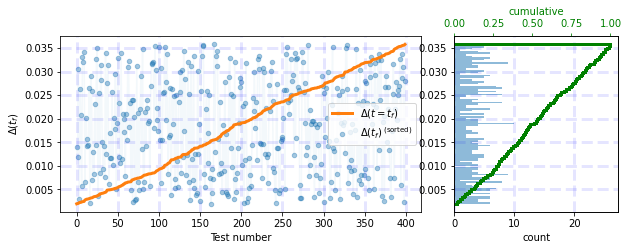
df = np.random.uniform(0.0359, 0.0018, 400)
test_histograms(df)
df = np.random.rayleigh(0.0359, 400)
test_histograms(df)
df = np.random.normal(0.0359, 0.0018, 400)
test_histograms(df)
Output before adding tight_layout:
Output after adding tight_layout:
Note:
You can use the w_pad argument of tight_layout to set the padding between subplots, for example using plt.tight_layout(w_pad=10) will result in the following: