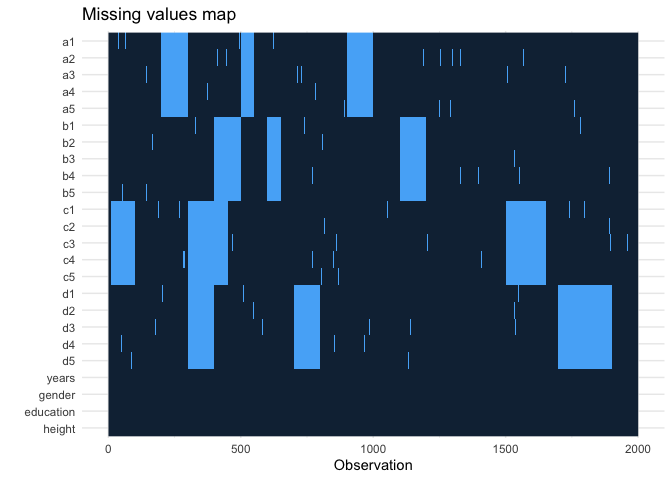
I have a 100 row, 200 variable dataset (mostly Likert-scale survey data as factors, but also some numeric data). I have attached a reprex with similar patterns of missing data below; some blank cells are sporadic, but for some rows, an entire scale is blank.
I wanted to use the mice package's multiple imputation before subscale calculation, but multiple imputation of a factor-heavy dataset is impractical. However, I need to address the sporadically-missing cells before calculating subscale totals because if one item in the scale is NA, then the subscale total will also be NA.
My goal is to conduct single/mean imputation on the sporadically-missing factor cells, then calculate subscale totals, and then conduct multiple imputation on the numeric subscale total variables, which will target the remaining large gaps.
I would like the sporadically-missing values to be imputed with the mean of the individual's remaining scale variables. E.g., if an individual missing df$c2 then that missing value would be imputed with the mean of c1, c3, c4, and c5.
How can I conduct single/mean imputation on a subset of data, dependent on the patterns of missingness within each row?
library(reprex)
library(tidyverse)
library(missMethods)
library(finalfit)
library(mice)
set.seed(1234)
a1 <- sample(1:3, 2000, replace=TRUE)
a2 <- sample(1:3, 2000, replace=TRUE)
a3 <- sample(1:3, 2000, replace=TRUE)
a4 <- sample(1:3, 2000, replace=TRUE)
a5 <- sample(1:3, 2000, replace=TRUE)
b1 <- sample(1:3, 2000, replace=TRUE)
b2 <- sample(1:3, 2000, replace=TRUE)
b3 <- sample(1:3, 2000, replace=TRUE)
b4 <- sample(1:3, 2000, replace=TRUE)
b5 <- sample(1:3, 2000, replace=TRUE)
c1 <- sample(1:3, 2000, replace=TRUE)
c2 <- sample(1:3, 2000, replace=TRUE)
c3 <- sample(1:3, 2000, replace=TRUE)
c4 <- sample(1:3, 2000, replace=TRUE)
c5 <- sample(1:3, 2000, replace=TRUE)
d1 <- sample(1:3, 2000, replace=TRUE)
d2 <- sample(1:3, 2000, replace=TRUE)
d3 <- sample(1:3, 2000, replace=TRUE)
d4 <- sample(1:3, 2000, replace=TRUE)
d5 <- sample(1:3, 2000, replace=TRUE)
years <- sample(18:70, 2000, replace=TRUE)
gender <- sample(c("male","female"), 2000, replace=TRUE, prob=c(0.5, 0.5))
education <- sample(c("highschool","college", "gradschool"), 2000, replace=TRUE, prob=c(1/3, 1/3, 1/3))
height <- sample(60:75, 2000, replace=TRUE)
df <- data.frame(a1, a2, a3, a4, a5,
b1, b2, b3, b4, b5,
c1, c2, c3, c4, c5,
d1, d2, d3, d4, d5,
years, gender, education, height)
facts <- df %>% select(contains("gender") | contains ("education")) %>% colnames()
cols <- df %>% select(ends_with("1") |
ends_with("2") |
ends_with("3") |
ends_with("4") |
ends_with("5")) %>% colnames()
df <- delete_MCAR(df, p = 0.01, cols_mis = cols) %>%
dplyr::mutate(across(all_of(facts), factor))
df[c(200:300, 500:550, 900:1000), 1:5] <- NA
df[c(400:500, 600:650, 1100:1200), 6:10] <- NA
df[c(10:100, 300:450, 1500:1650), 11:15] <- NA
df[c(300:400, 700:800, 1700:1900), 16:20] <- NA
## I think mean imputation of the sporadically-missing cells would occur here
missing_plot(df)
df <- df %>%
rowwise() %>%
mutate(a_mean = mean(c(a1, a2, a3, a4, a5))) %>%
mutate(b_mean = mean(c(b1, b2, b3, b4, b5))) %>%
mutate(c_mean = mean(c(c1, c2, c3, c4, c5))) %>%
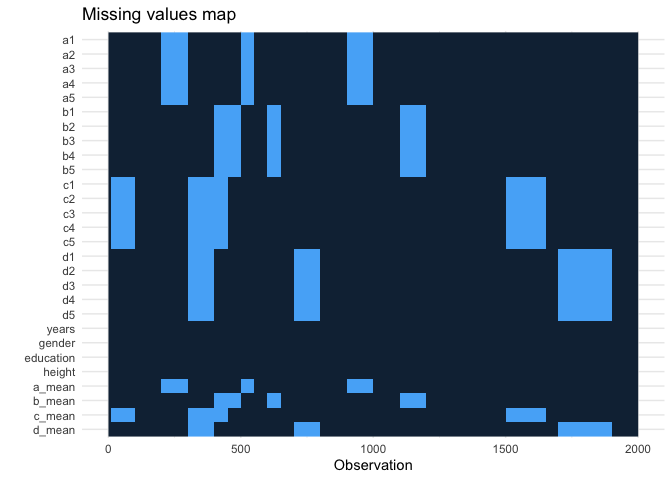
mutate(d_mean = mean(c(d1, d2, d3, d4, d5)))
df <- df %>%
select(ends_with("mean") | contains("years") | contains("gender") | contains("education") | contains("height"))
imp_df <- mice::mice(df, m = 5, print = FALSE)
com <- mice::complete(imp_df)
CodePudding user response:
Am I understanding right that you just want to take the mean for each a1 ... d5 individually, only where the missing values are sporadically missing? Then we can define a function that sifts through to identify consecutive NA values that are less than a certain length. I based the method on 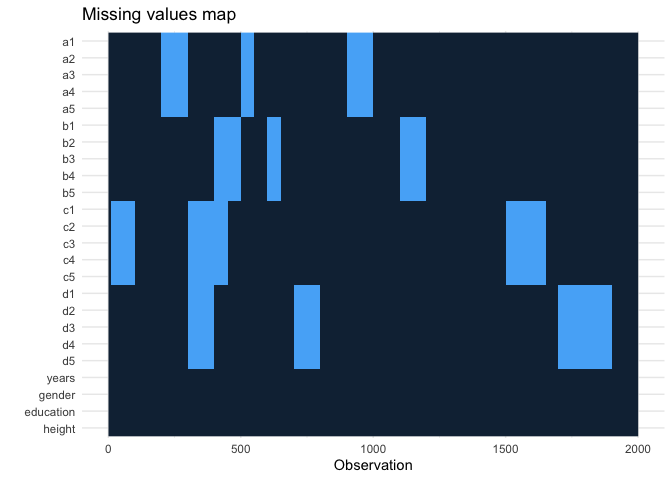
Infill with mean of other scale variables
If we want to infill with other scale variables, we take a similar approach. Just a couple notes:
- We need to remove missing values from the
rowwise()scale means, because otherwise it'sNAwherever we have a missing value and we can't do any imputation anyway. - We should always
ungroup()after usingrowwise()otherwise it's easy to forget and return strange results. - We'll use some of the functionality of
across()to accesscur_column()name, subset the letter, and get the relatedx_meancolumn in.data. - Define an
infill_fun()to do the above wrappingna_search().
df <- df %>%
rowwise() %>%
mutate(a_mean = mean(c(a1, a2, a3, a4, a5), na.rm = T)) %>%
mutate(b_mean = mean(c(b1, b2, b3, b4, b5), na.rm = T)) %>%
mutate(c_mean = mean(c(c1, c2, c3, c4, c5), na.rm = T)) %>%
mutate(d_mean = mean(c(d1, d2, d3, d4, d5), na.rm = T)) %>%
ungroup()
infill_fun <- function(x, threshold, df, cur_col) {
mean_col <- paste0(substr(cur_col, 1, 1), "_mean")
ifelse(na_search(x, threshold),
df[[mean_col]],
x)
}
df %>%
mutate(across(
matches("[a-z][1-9]"),
~ infill_fun(
.x,
threshold = 5,
df = .data,
cur_col = cur_column()
)
)) %>%
missing_plot()