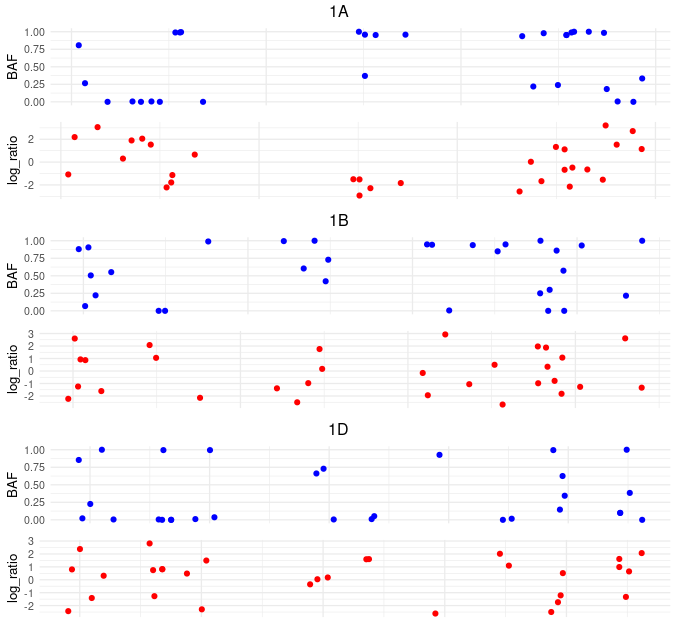
I have a data like this
structure(list(Chr_id = c("1A", "1A", "1A", "1A", "1A", "1A",
"1A", "1A", "1A", "1A", "1A", "1A", "1A", "1A", "1A", "1A", "1A",
"1A", "1A", "1A", "1A", "1A", "1A", "1A", "1A", "1A", "1A", "1A",
"1A", "1A", "1B", "1B", "1B", "1B", "1B", "1B", "1B", "1B", "1B",
"1B", "1B", "1B", "1B", "1B", "1B", "1B", "1B", "1B", "1B", "1B",
"1B", "1B", "1B", "1B", "1B", "1B", "1B", "1B", "1B", "1B", "1D",
"1D", "1D", "1D", "1D", "1D", "1D", "1D", "1D", "1D", "1D", "1D",
"1D", "1D", "1D", "1D", "1D", "1D", "1D", "1D", "1D", "1D", "1D",
"1D", "1D", "1D", "1D", "1D", "1D", "1D"), Start = c(111381744,
312370649, 106700529, 577069227, 13903751, 484952182, 546871249,
549719683, 560876463, 343004004, 301369910, 295169503, 82127411,
7487575, 531277958, 499574273, 508318998, 112534618, 135111292,
71379690, 301371731, 62654363, 474295484, 462885209, 90752869,
37060118, 516181841, 586104884, 508318998, 513528750, 1205670212,
1259468661, 633852277, 594372162, 1183474873, 843578162, 1017681165,
699258349, 1073242080, 1279167768, 880972535, 867779030, 1155145280,
608978435, 1044603066, 1023765196, 751735249, 1164927036, 614786148,
897594687, 1113070984, 1184408414, 894457006, 1103476280, 606094615,
1175159133, 1166717363, 691495384, 1155547991, 602113219, 1309902292,
1367857833, 1652686374, 1367857833, 1300204987, 1290581423, 1357433559,
1751459106, 1748871019, 1592296298, 1361382337, 1388111038, 1695238483,
1535545547, 1761828229, 1697034092, 1360290688, 1319668145, 1495335256,
1687491614, 1400339618, 1645319622, 1503836513, 1537716335, 1743422939,
1404043162, 1743422939, 1489337730, 1693017288, 1293616598),
BAF = c(0.989473684210526, 0.953125, 0.989583333333333, 0,
0.265625, 0.979166666666667, 0.984375, 0.182291666666667,
0.00520833333333333, 0.958333333333333, 0.958333333333333,
1, 0.00520833333333333, 0.807291666666667, 1, 0.239583333333333,
0.953125, 0.994791666666667, 0, 0, 0.369791666666667, 0.00520833333333333,
0.21875, 0.936842105263158, 0, 0, 1, 0.333333333333333, 0.953125,
0.989583333333333, 0.932291666666667, 0.215789473684211,
0.552083333333333, 0.880208333333333, 0.572916666666667,
0.994736842105263, 0.947916666666667, 0, 0.9375, 1, 1, 0.604166666666667,
0.25, 0.505208333333333, 0.00520833333333333, 0.942708333333333,
0.989583333333333, 0, 0.221052631578947, 0.729166666666667,
0.947916666666667, 0, 0.421875, 0.848958333333333, 0.905263157894737,
0.859375, 0.3, 0, 1, 0.0677083333333333, 1, 0, 0.0157894736842105,
0, 0.226315789473684, 0.854166666666667, 0.00526315789473684,
0.384210526315789, 1, 0.927083333333333, 0.994791666666667,
0.0105263157894737, 0.625, 0.0104166666666667, 0, 0.34375,
0, 0.00520833333333333, 0.729166666666667, 0.994736842105263,
0.994791666666667, 0, 0.00520833333333333, 0.0520833333333333,
0.0989583333333333, 0.0364583333333333, 0.0989583333333333,
0.661458333333333, 0.145833333333333, 0.0210526315789474),
log_ratio = c(-1.785, -2.286, -2.218, 2.715, 2.189, -1.671,
-1.541, 3.209, 1.528, -1.836, -1.516, -1.501, 2.051, -1.082,
-0.648, 1.326, 1.114, -1.137, 0.655, 1.895, -2.928, 0.306,
0.028, -2.574, 1.532, 3.061, -0.482, 1.142, -0.673, -2.15,
-1.268, 2.613, -1.607, -2.225, -1.82, -1.386, -0.157, 1.054,
-1.055, -1.332, -0.971, -2.503, 1.965, 0.928, 2.926, -1.94,
-2.144, 1.876, 0.87, 0.171, -2.682, 1.073, 1.758, 0.499,
-1.241, -0.788, 0.342, 2.076, -0.98, 2.599, -1.413, 0.813,
1.095, 0.822, 2.381, -2.431, 2.814, 0.646, -1.328, -2.616,
-1.27, 0.481, -1.208, 1.588, 2.067, 0.514, 0.747, 0.312,
0.04, -2.497, -2.291, 2.014, 0.177, 1.6, 0.986, 1.49, 1.613,
-0.354, -1.739, 0.801)), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -90L))
And I use this code
ggplot(data_sample, aes(x = Start))
geom_point(aes(y = BAF),
color = 'blue')
geom_point(aes(y = log_ratio),
color = 'red')
theme_minimal(base_size = 20)
facet_wrap(~ Chr_id,
ncol = 1,
nrow = 3,
scales = 'free')
theme(axis.title.x = element_blank(),
axis.text.x = element_blank(),
legend.position = 'none')
to create plot like this one
Red points have very different scale from blue ones and I tried to create a pairs of plots for each group by Chr_id (1A, 1B, 1D) values and to move geom_point contains red points on the plot below the plot with blue points. I have no idea is there even possible because I already use facet_wrap and I need to make like subgrid for every Chr_id plot produced by facet_wrap. I think it is possible using grid.arrange function but in real data Chr_id contains 17 categories and code will be too bulky. Is there more elegant solution for this? Thanks.
Update 1
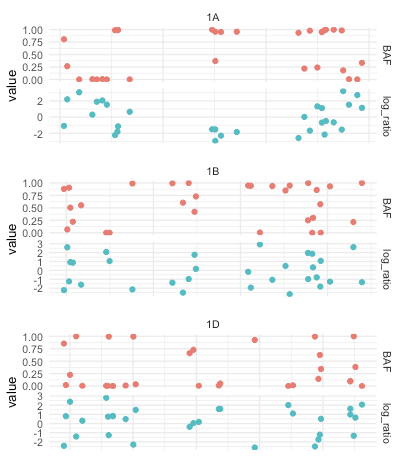
I want to do plot like this
I did this using grid.arange and code is too big as I mentioned before. Also x-axis for pairs of subplots is not synchronized.
CodePudding user response:
If you make a list with all your plots, you can use do.call to call grid.arrange on the whole list. It's easiest to make a list of plots if we write a little function to produce 1 plot. I changed your base_size to 10 to make the image fit better on Stack Overflow--you can of course change it back and customize the one_plot function however you'd like.
library(dplyr)
library(tidyr)
long_data = data_sample %>%
pivot_longer(cols = c("BAF", "log_ratio")) %>%
mutate(grouper = paste(Chr_id, name, sep = ":"))
one_plot = function(chr, data) {
ggplot(filter(long_data, Chr_id == chr), aes(x = Start, color = name, y = value))
geom_point()
theme_minimal(base_size = 20)
facet_grid(rows = vars(name),
cols = vars(Chr_id),
scales = 'free')
theme(axis.title.x = element_blank(),
axis.text.x = element_blank(),
legend.position = 'none')
}
plot_list = lapply(unique(long_data$Chr_id), one_plot, data = long_data)
do.call(grid.arrange, c(plot_list, ncol = 1))