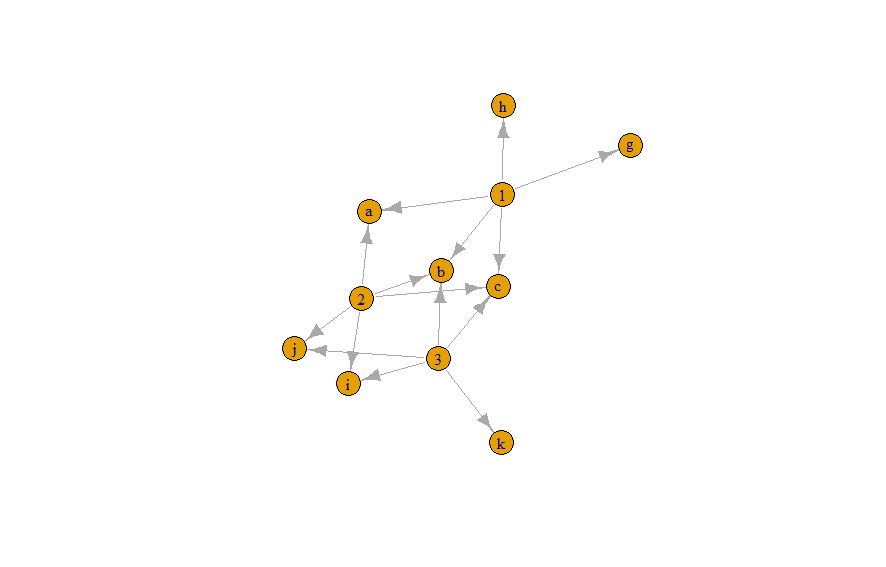
I am trying to find the number of shared and unique features between several clusters (nodes) and visualize them. The data I have is in 2 columns. I think I need to get it into target/source format but I cannot figure it out from this current format.
Here is some example data:
df <- data.frame(cluster = c(rep(1, 5), rep(2, 5), rep(3, 5)),
feature = c(letters[1:3], letters[7:8], letters[1:3], letters[9:10], letters[2:3], letters[9:11]))
> df
cluster feature
1 1 a
2 1 b
3 1 c
4 1 g
5 1 h
6 2 a
7 2 b
8 2 c
9 2 i
10 2 j
11 3 b
12 3 c
13 3 i
14 3 j
15 3 k
I want to show that cluster 1 shares a with 1 other cluster, cluster 1 shares b with 2 other clusters, cluster 2 shares i with cluster 3 etc.
I have tried so many combinations of tidyr, plyr, dplyr code but I can't figure it out. For example this basic code gives me the number of shared partners between clusters, but not which partner it is shared with.
df2 <- df %>%
group_by(feature) %>%
mutate(n_gene = n())
> df2
# A tibble: 15 × 3
# Groups: feature [8]
cluster feature n_gene
<dbl> <chr> <int>
1 1 a 2
2 1 b 3
3 1 c 3
4 1 g 1
5 1 h 1
6 2 a 2
7 2 b 3
8 2 c 3
9 2 i 2
10 2 j 2
11 3 b 3
12 3 c 3
13 3 i 2
14 3 j 2
15 3 k 1
My goal is to have either a network like something below (credit to 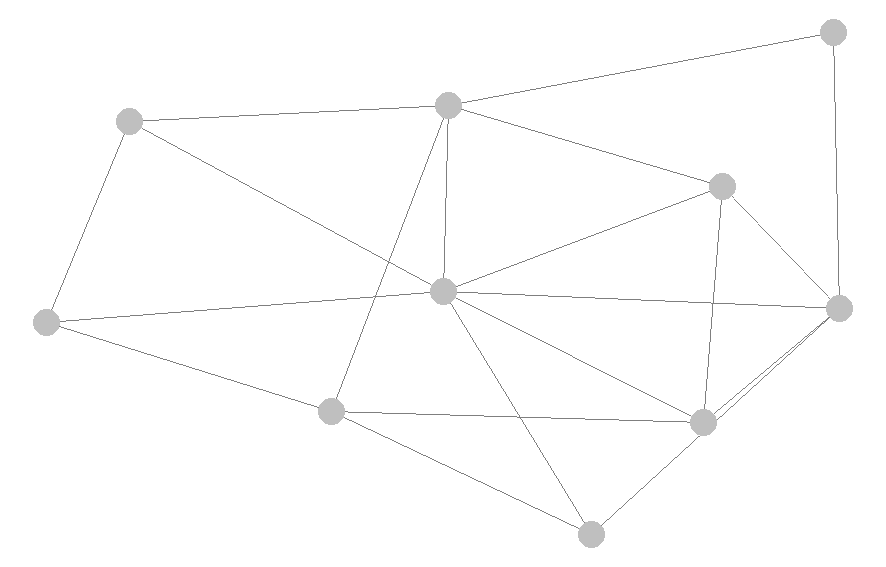
I tried plotting your graph using iagraph as well. What it does is, create a node between a cluster and a feature that is wrong but still, I'll include it to show you what's wrong.
library(igraph)
df.g <- graph.data.frame(d = df, directed = TRUE)
df.g
plot(df.g, vertex.label = V(df.g)$name)
CodePudding user response:
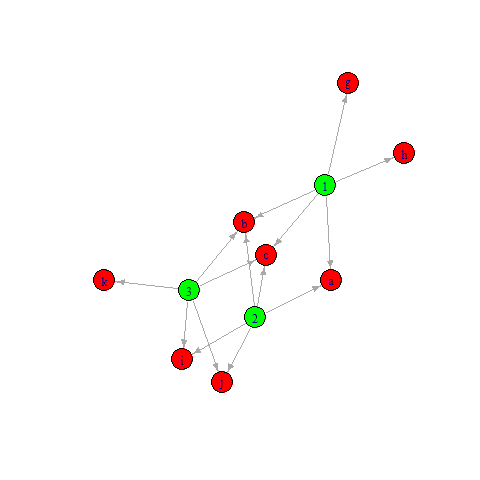
Maybe something like this?
df %>%
graph_from_data_frame() %>%
set_vertex_attr(
name = "color",
value = c("green", "red")[1 (names(V(.)) %in% df$feature)]
) %>%
plot()