I'm using Matplotlib's imshow to plot data as a function of two variables, one of which is time. There were significant interruptions in data collection in many cases, and I want to note that on the graph using whitespace (or something similar) separating data that has a lapse in between them. I'm having difficulty putting this into practice, however. I inserted a dummy line of data consisting of NaN values, which plot as white on a color map. The issue is in the visual appearance. I'm using the catrom interpolation to create the heatmap, which causes a wider whitespace than I'd prefer. Using another interpolation scheme doesn't produce plots I'm happy with.
I can't provide a MWE because my data is sufficiently large that I can't supply it, and I don't have the time to create a dummy data set that would show this well enough. However, I can provide the code I use to create the plots themselves and their output. Here is the code to produce something similar to what I'd like to see:
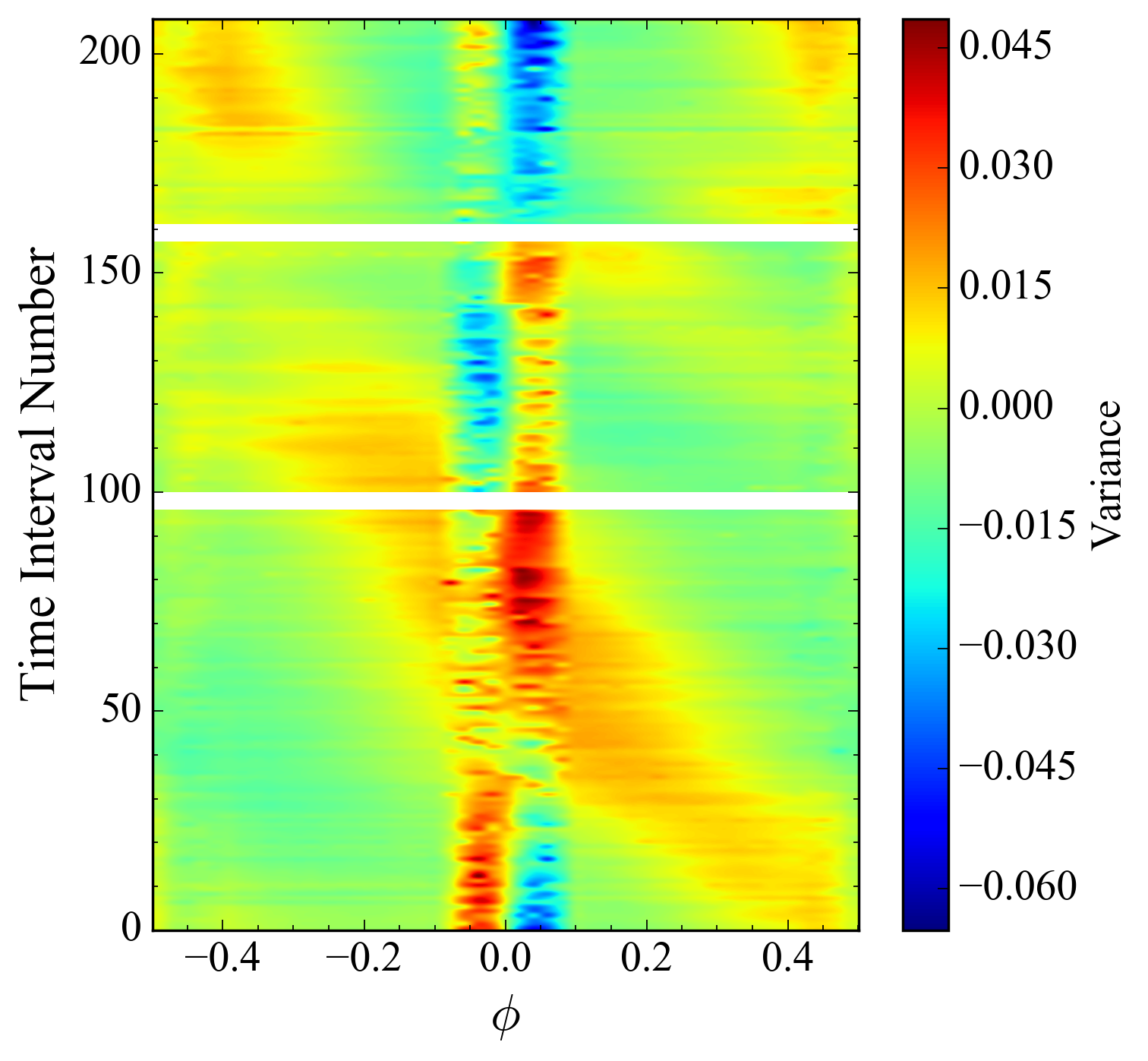
# Plot the time and phase difference as a color map showing variation over both domains.
img = ax2D.imshow(variance_array, extent=[-0.5, 0.5, float(period_numbers[-1]), 0.0], aspect="auto",
interpolation="nearest")
plt.colorbar(img, ax=ax2D, label="Variance", spacing="proportional")
ax2D.minorticks_on()
ax2D.invert_yaxis()
This results in:
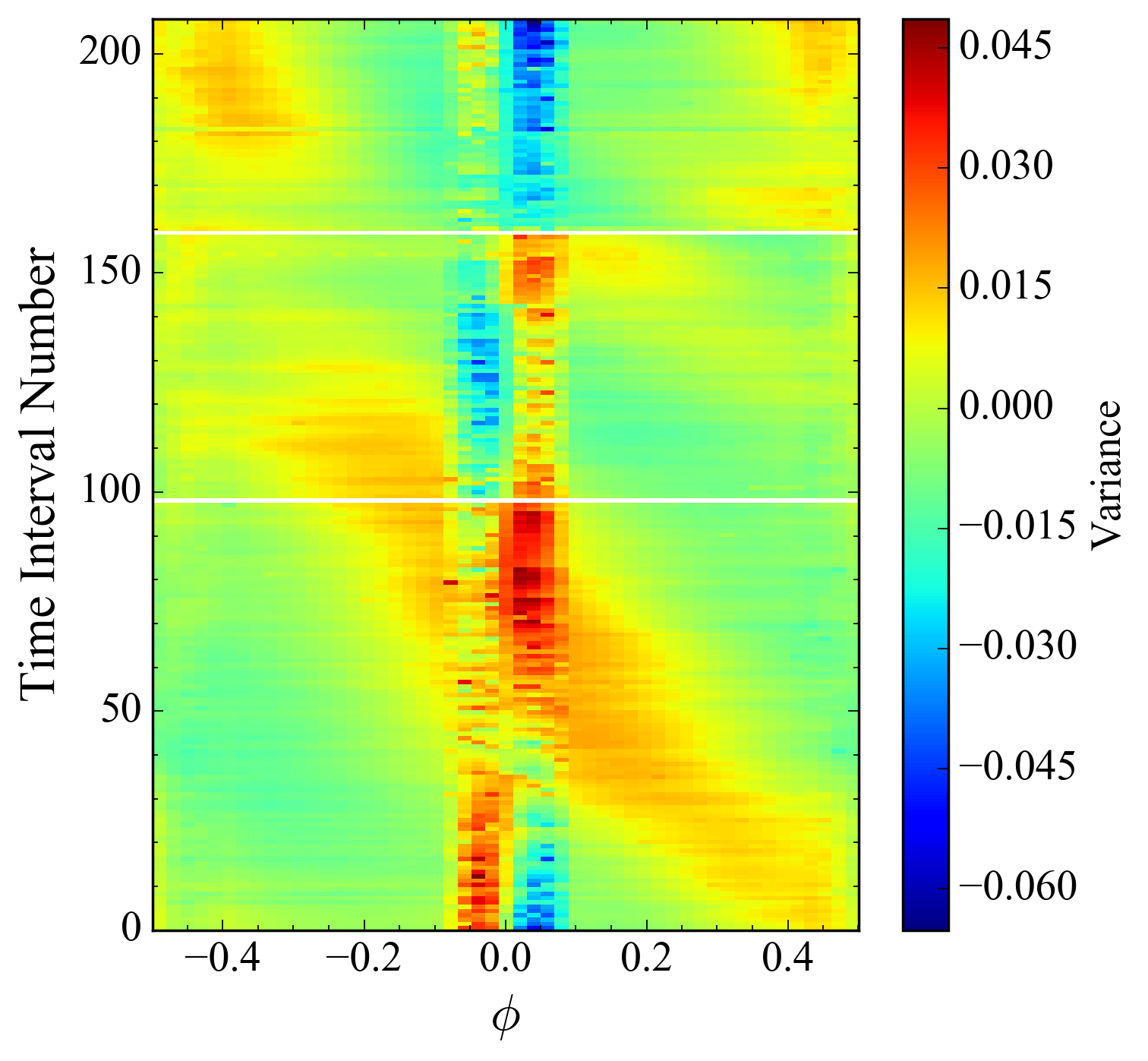
However, it's so pixelated! That's not the quality of something I'd like to publish in a paper, to say nothing of my dissertation. The code I have been using to produce these images previously is:
# Plot the time and phase difference as a color map showing variation over both domains.
img = ax2D.imshow(variance_array, extent=[-0.5, 0.5, float(period_numbers[-1]), 0.0], aspect="auto",
interpolation="catrom")
plt.colorbar(img, ax=ax2D, label="Variance", spacing="proportional")
ax2D.minorticks_on()
ax2D.invert_yaxis()
This results in:
It's much smoother, but look at how wide the whitespace is! It makes it harder to tell any trends that occurred over the lapsed data, and it actually removes some useful data.
What's the best way to indicate this sort of break in the data? Am I going about it the best way, or is there something better I haven't considered? If this is the best method, how do I fix how wide the whitespace is?
CodePudding user response:
For matplotlib >= 3.5.0
If you have matplotlib 3.5.0 or higher, you can use the imshow parameter 
For matplotlib < 3.5.0
One hack that you could try is to apply a fake mask over your data after the interpolation by using 1. a second call to imshow and 2. a colormap of only white values.
Here's an example:
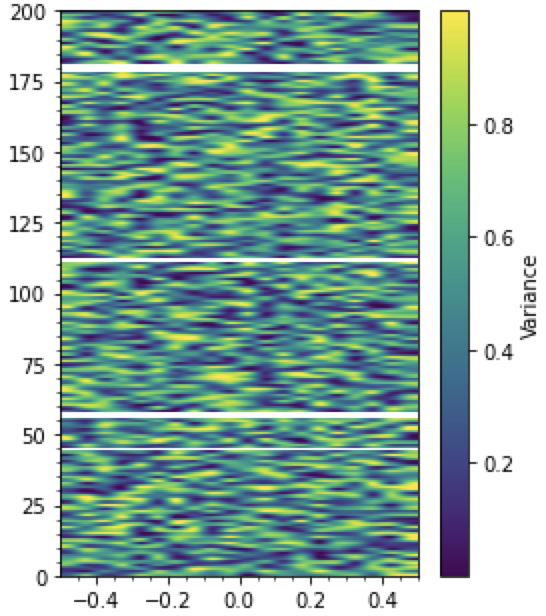
import matplotlib
import matplotlib.pyplot as plt
# Plot the time and phase difference as a color map showing variation over both domains.
fig, ax2D = plt.subplots(figsize=plt.figaspect(1.3))
variance_array = np.random.rand(180, 20)
img = ax2D.imshow(variance_array, extent=[-0.5, 0.5, 200, 0.0], aspect="auto",
interpolation="catrom", alpha=1)
## Mask broken timepoints
# (generate random broken_timepoints)
broken_timepoints = [40, 50, 51, 100, *list(range(160, 163))]
mask = np.full_like(variance_array, np.nan)
mask[broken_timepoints, :] = 0
# (apply white mask)
white_only_cmap = matplotlib.colors.ListedColormap(["white"])
_ = ax2D.imshow(mask, extent=[-0.5, 0.5, 200, 0.0], aspect="auto",
interpolation="nearest", alpha=1, cmap=white_only_cmap)
plt.colorbar(img, ax=ax2D, label="Variance", spacing="proportional")
ax2D.minorticks_on()
ax2D.invert_yaxis()