I have defined a custom function and tested the function to make sure that it works but I am unable to apply it to a list in order to obtain a distance matrix.
The code I have is:
library(Biostrings)
library(proxy)
#import the sequences using Biostrings
indf<-readAAStringSet("C:/Users/jamie/OneDrive/Documents/Junk/SAMPLEFASTA.fasta")
#Assign the names and sequences to different variables
seqAAname<-names(indf)
seqz<-paste(indf)
#Put just the sequences into a dataframe
indf2<-data.frame(seqz)
#Convert the sequences into a list
indf3<-as.list(indf2)
#Define a custom function to return the alignment score between two sequences (pairwise)
customalnfunc <- function(X, Y){
pairwiseAlignment(X, Y,
substitutionMatrix = "BLOSUM45", gapOpening = 1, gapExtension = 3)
}
#Test the function but not as a function (This works fine)
testfreefunc<- pairwiseAlignment(AAString("PEHQRSTVE"),AAString("PQHQRETVE"),
substitutionMatrix = "BLOSUM45", gapOpening = 1, gapExtension = 3)
print(testfreefunc@score)
#Test the function as a fucntion to make sure it works (This works fine)
testfuncout <- customalnfunc(AAString("PEHQRSTVE"),AAString("PQHQRETVE"))
print(testfuncout@score)
#Apply the custom function to all possible pairs using proxy::dist with the custom function (This does not work, it returns 0)
outalnmatrix <- proxy::dist(indf3, method = customalnfunc)
outalnmatrix
The SAMPLEFASTA.fasta file contains:
>SeqA
PEHQRSTVE
>SeqB
PQHQRETVE
>SeqC
RQHERSEVE
The desired output from outalnmatrix is:
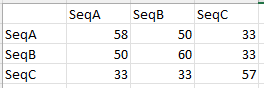
I have tried passing the input data to proxy::dist as a list and a matrix.
How can I make this work?
CodePudding user response:
You don't need to use the proxy package as proxy::dist is meant to icompare rows of matrix/dataframes against each other. Since you want to compare strings, you can use outer. However, you need to tweak your customalnfunc function, so that it returns only a number (scoreOnly = TRUE).
library(Biostrings)
seqz <- c("PEHQRSTVE", "PQHQRETVE", "RQHERSEVE")
customalnfunc <- function(X, Y){
pairwiseAlignment(X, Y,
substitutionMatrix = "BLOSUM45",
gapOpening = 1,
gapExtension = 3,
scoreOnly = TRUE)
}
outer(seqz, seqz, customalnfunc)
#>
[,1] [,2] [,3]
[1,] 58 50 33
[2,] 50 60 33
[3,] 33 33 57
